2024 年 4 巻 p. 117-125
2024 年 4 巻 p. 117-125
The peak separation of all 136 tetra- to octa-chlorinated dibenzo-p-dioxins and dibenzofurans on two types of 50% phenyl methyl siloxane gas chromatography columns, DB-17ms and VF-17ms, is presented. Four congeners of the seventeen 2,3,7,8-congeners, 2,3,7,8-TeCDD, 1,2,3,7,8-PeCDD, 2,3,7,8-TeCDF, and 1,2,3,4,7,8-HxCDF, were not separated clearly on DB-17ms. On VF-17ms, 2,3,7,8-TeCDD, 2,3,7,8-TeCDF, 1,2,3,7,8-PeCDF, and 1,2,3,6,7,8-HxCDF were not clearly separated. The other 2,3,7,8-congeners showed good separation on both columns. Therefore, the concentrations of all seventeen 2,3,7,8-congeners can be obtained by combining the measurement results from DB-17ms or VF-17ms with those from DB-5ms or similar columns.

Polychlorinated dibenzo-p-dioxins (PCDDs) and polychlorinated dibenzofurans (PCDFs), referred to as dioxins, are toxic compounds that are highly persistent in the environment. Up to eight chlorines can be substituted in the basic structure of dioxins; there are 210 dioxin congeners (75 PCDDs and 135 PCDFs), which vary with the number and position of chlorine substitutions. Of these 210 congeners, 136 (49 PCDDs and 87 PCDFs) with four or more chlorine atoms, including seventeen 2,3,7,8-chlorine-substituted congeners, are subject to environmental and industrial emissions monitoring. The toxicity equivalency, which is calculated from the concentrations of the seventeen 2,3,7,8-congeners and the toxicity equivalency factor values assigned to each, is usually used to assess the overall toxicity of dioxins (Van den Berg et al., 2006). The concentrations of the seventeen 2,3,7,8-congeners must be accurately measured to calculate an accurate toxic equivalency. Dioxins are usually quantified by using gas chromatography-mass spectrometry (GC-MS) with a capillary column. Separation of the dioxin congeners on GC is of interest to analysts.
Many researchers have reported the separation of dioxin congeners on various capillary columns (Ryan et al., 1991; Bacher and Ballschmiter, 1992; Fraisse et al., 1994; Abad et al., 1997; Matsumura et al., 2003; Fishman et al., 2004, 2007, 2011; Do et al., 2013; Iwakiri and Enomoto, 2018; Stultz and Dorman, 2020). In particular, reports on the separation of all 136 tetra- to octa-chlorinated congeners (Ryan et al., 1991; Bacher and Ballschmiter, 1992; Matsumura et al., 2003; Fishman et al., 2011; Do et al., 2013) are helpful for accurate peak assignment. However, currently, no capillary column capable of separating all seventeen 2,3,7,8-congeners has been reported. Therefore, multiple measurements on different columns are needed to separate all seventeen. Identifying peak assignments on various capillary columns provides analysts with more options. Some studies have conducted the full assignment of the 136 congeners on “DB-5” (5% phenyl methyl silicone or similar)-type columns (Ryan et al., 1991; Fishman et al., 2011), but there have been few on the “DB-17” (50% phenyl methyl silicone or similar) type (Ryan et al., 1991). Therefore, here, we report the confirmed peak separation of all 136 tetra- to octa-dioxins by using two commercially available “50% phenyl”-type columns, namely DB-17ms and VF-17ms. To our knowledge, full assignment of the 136 dioxin congeners by using these two columns has not been reported before.
The abbreviations for the PCDD/PCDF congeners were as follows: CDD=chlorinated dibenzo-p-dioxin; CDF=chlorinated dibenzofuran; Te=tetra; Pe=penta; Hx=hexa; Hp=hepta; O=octa.
A Comprehensive Polychlorinated Dioxin and Furan Column Defining Kit (Cambridge Isotope Laboratories (CIL), Andover, MA, USA) was used to determine the retention order of all 136 tetra- to octa-CDDs/CDFs. The standard solution was prepared into 38 mixtures (nos. 1 to 38) containing three to four congeners with no overlapping homologs. 13C12-1,2,7,8-TeCDF (Wellington Laboratories, Guelph, Ontario, Canada) was added to the mixtures to allow compensation for shifts in retention time (RT) that may have occurred during the analysis. The concentration of each congener was approximately 5 pg/μL in nonane. In addition, mixed solutions for each homolog (nos. 39 to 42) were prepared. The concentration of each congener was approximately 0.6 pg/μL in nonane. The combinations of congeners in each standard mixture are shown in Table 1.
| Mixture No. | Congeners | ||||
|---|---|---|---|---|---|
| 1 | 1,2,3,4-TeCDF | 1,2,3,4,6-PeCDF | 1,2,3,4,6,7-HxCDF | 1,2,3,4-TeCDD | 13C12-1,2,7,8-TeCDF |
| 2 | 1,2,3,6-TeCDF | 1,2,3,4,7-PeCDF | 1,2,3,4,6,8-HxCDF | 1,2,3,6-TeCDD | 13C12-1,2,7,8-TeCDF |
| 3 | 1,2,3,7-TeCDF | 1,2,3,4,8-PeCDF | 1,2,3,4,6,9-HxCDF | 1,2,3,7-TeCDD | 13C12-1,2,7,8-TeCDF |
| 4 | 1,2,3,8-TeCDF | 1,2,3,4,9-PeCDF | 1,2,3,4,7,8-HxCDF | 1,2,3,8-TeCDD | 13C12-1,2,7,8-TeCDF |
| 5 | 1,2,3,9-TeCDF | 1,2,3,6,7-PeCDF | 1,2,3,4,7,9-HxCDF | 1,2,3,9-TeCDD | 13C12-1,2,7,8-TeCDF |
| 6 | 1,2,4,6-TeCDF | 1,2,3,6,8-PeCDF | 1,2,3,4,8,9-HxCDF | 1,2,4,6-TeCDD | 13C12-1,2,7,8-TeCDF |
| 7 | 1,2,4,7-TeCDF | 1,2,3,6,9-PeCDF | 1,2,3,6,7,8-HxCDF | 1,2,4,7-TeCDD | 13C12-1,2,7,8-TeCDF |
| 8 | 1,2,4,8-TeCDF | 1,2,3,7,8-PeCDF | 1,2,3,6,7,9-HxCDF | 1,2,4,8-TeCDD | 13C12-1,2,7,8-TeCDF |
| 9 | 1,2,4,9-TeCDF | 1,2,3,7,9-PeCDF | 1,2,3,6,8,9-HxCDF | 1,2,4,9-TeCDD | 13C12-1,2,7,8-TeCDF |
| 10 | 1,2,6,7-TeCDF | 1,2,3,8,9-PeCDF | 1,2,3,7,8,9-HxCDF | 1,2,6,7-TeCDD | 13C12-1,2,7,8-TeCDF |
| 11 | 1,2,6,8-TeCDF | 1,2,4,6,7-PeCDF | 1,2,4,6,7,8-HxCDF | 1,2,6,8-TeCDD | 13C12-1,2,7,8-TeCDF |
| 12 | 1,2,6,9-TeCDF | 1,2,4,6,8-PeCDF | 1,2,4,6,7,9-HxCDF | 1,2,6,9-TeCDD | 13C12-1,2,7,8-TeCDF |
| 13 | 1,2,7,8-TeCDF | 1,2,4,6,9-PeCDF | 1,2,4,6,8,9-HxCDF | 1,2,7,8-TeCDD | 13C12-1,2,7,8-TeCDF |
| 14 | 1,2,7,9-TeCDF | 1,2,4,7,8-PeCDF | 1,3,4,6,7,8-HxCDF | 1,2,7,9-TeCDD | 13C12-1,2,7,8-TeCDF |
| 15 | 1,2,8,9-TeCDF | 1,2,4,7,9-PeCDF | 1,3,4,6,7,9-HxCDF | 1,2,8,9-TeCDD | 13C12-1,2,7,8-TeCDF |
| 16 | 1,3,4,6-TeCDF | 1,2,4,8,9-PeCDF | 2,3,4,6,7,8-HxCDF | 1,3,6,8-TeCDD | 13C12-1,2,7,8-TeCDF |
| 17 | 1,3,4,7-TeCDF | 1,2,6,7,8-PeCDF | 1,2,3,4,6-PeCDD | 1,3,6,9-TeCDD | 13C12-1,2,7,8-TeCDF |
| 18 | 1,3,4,8-TeCDF | 1,2,6,7,9-PeCDF | 1,2,3,4,7-PeCDD | 1,3,7,8-TeCDD | 13C12-1,2,7,8-TeCDF |
| 19 | 1,3,4,9-TeCDF | 1,3,4,6,7-PeCDF | 1,2,3,6,7-PeCDD | 1,3,7,9-TeCDD | 13C12-1,2,7,8-TeCDF |
| 20 | 1,3,6,7-TeCDF | 1,3,4,6,8-PeCDF | 1,2,3,6,8-PeCDD | 1,4,6,9-TeCDD | 13C12-1,2,7,8-TeCDF |
| 21 | 1,3,6,8-TeCDF | 1,3,4,6,9-PeCDF | 1,2,3,6,9-PeCDD | 1,4,7,8-TeCDD | 13C12-1,2,7,8-TeCDF |
| 22 | 1,3,6,9-TeCDF | 1,3,4,7,8-PeCDF | 1,2,3,7,8-PeCDD | 2,3,7,8-TeCDD | 13C12-1,2,7,8-TeCDF |
| 23 | 1,3,7,8-TeCDF | 1,3,4,7,9-PeCDF | 1,2,3,7,9-PeCDD | 13C12-1,2,7,8-TeCDF | |
| 24 | 1,3,7,9-TeCDF | 1,3,6,7,8-PeCDF | 1,2,3,8,9-PeCDD | 13C12-1,2,7,8-TeCDF | |
| 25 | 1,4,6,7-TeCDF | 1,4,6,7,8-PeCDF | 1,2,4,6,7-PeCDD | 13C12-1,2,7,8-TeCDF | |
| 26 | 1,4,6,8-TeCDF | 2,3,4,6,7-PeCDF | 1,2,4,6,8-PeCDD | 13C12-1,2,7,8-TeCDF | |
| 27 | 1,4,6,9-TeCDF | 2,3,4,6,8-PeCDF | 1,2,4,6,9-PeCDD | 13C12-1,2,7,8-TeCDF | |
| 28 | 1,4,7,8-TeCDF | 2,3,4,7,8-PeCDF | 1,2,4,7,8-PeCDD | 13C12-1,2,7,8-TeCDF | |
| 29 | 1,6,7,8-TeCDF | 1,2,3,4,6,7-HxCDD | 1,2,4,7,9-PeCDD | 13C12-1,2,7,8-TeCDF | |
| 30 | 2,3,4,6-TeCDF | 1,2,3,4,6,8-HxCDD | 1,2,4,8,9-PeCDD | 13C12-1,2,7,8-TeCDF | |
| 31 | 2,3,4,7-TeCDF | 1,2,3,4,6,9-HxCDD | 1,2,3,4,6,7,8-HpCDF | 13C12-1,2,7,8-TeCDF | |
| 32 | 2,3,4,8-TeCDF | 1,2,3,4,7,8-HxCDD | 1,2,3,4,6,7,9-HpCDF | 13C12-1,2,7,8-TeCDF | |
| 33 | 2,3,6,7-TeCDF | 1,2,3,6,7,8-HxCDD | 1,2,3,4,6,8,9-HpCDF | 13C12-1,2,7,8-TeCDF | |
| 34 | 2,3,6,8-TeCDF | 1,2,3,6,7,9-HxCDD | 1,2,3,4,7,8,9-HpCDF | 13C12-1,2,7,8-TeCDF | |
| 35 | 2,3,7,8-TeCDF | 1,2,3,6,8,9-HxCDD | OCDF | 13C12-1,2,7,8-TeCDF | |
| 36 | 2,4,6,7-TeCDF | 1,2,3,7,8,9-HxCDD | 1,2,3,4,6,7,8-HpCDD | 13C12-1,2,7,8-TeCDF | |
| 37 | 2,4,6,8-TeCDF | 1,2,4,6,7,9-HxCDD | 1,2,3,4,6,7,9-HpCDD | 13C12-1,2,7,8-TeCDF | |
| 38 | 3,4,6,7-TeCDF | 1,2,4,6,8,9-HxCDD | OCDD | 13C12-1,2,7,8-TeCDF | |
| 39 | TeCDFs, | ||||
| 40 | PeCDFs | HxCDDs | |||
| 41 | HxCDFs | PeCDDs | Hp–OCDFs | Hp–OCDDs | |
| 42 | TeCDDs | ||||
Two chromatographic columns (length: 60 m; id: 0.25 mm; film thickness: 0.25 μm), namely DB-17ms (Agilent Technologies, Santa Clara, CA, USA) and VF-17ms (Agilent Technologies), were tested for the separation of PCDDs/PCDFs. Both columns are 50%-phenyl-type columns, but the substitution position of the phenyl group in the liquid phase differs, with silphenylene–dimethylsiloxane for DB-17ms and diphenylsiloxane–dimethylsiloxane for VF-17ms (Fig. 1).

PCDDs/PCDFs in the standard solutions were measured by using high-resolution GC-MS (Agilent 7890A GC (Agilent Technologies)+JMS-800D (JEOL, Tokyo, Japan)). MS was performed under positive electron ionization (resolution: >10,000), and data were acquired in selected ion monitoring mode. The same GC temperature program was used for the two columns: 130°C (2 min) to 210°C (15°C/min) to 310°C (3°C/min), and finally to 320°C (5°C/min, 17 min) (total: 59.7 min). Injection was performed in splitless mode at a temperature of 280°C. Helium was used as the carrier gas for the column, and the flow rate was 1.5 mL/min. The monitoring ion channels (m/z) were 321.8936 for TeCDDs; 355.8546 for PeCDDs; 389.8156 for HxCDDs; 423.7767 for HxCDDs; 459.7348 for OCDD; 305.8987 for TeCDFs; 339.8597 for PeCDFs; 373.8207 for HxCDFs; 407.7818 for HpCDFs; 443.7398 for OCDF; and 317.9389 for 13C12-1278-TeCDF. The data acquisition time of each ion channel was 27 ms and the cycle time was 500 ms. Increasing the number of monitoring channels reduces the data acquisition time, resulting in reduced sensitivity. To confirm peak separation more clearly, the number of monitoring channels per homolog was set to one, and the number of data points in the chromatogram was increased. Although analytical manuals in Japan require the monitoring of two or more homolog channels for the determination of dioxins (JSA, 2020a, 2020b; MOE, 2022a, 2022b, 2022c), one monitoring channel was considered sufficient because each congener of the standard material used was prepared with high purity.
Peak assignment was performed by using the dioxin quantitative program DioK ver. 4.02 (JEOL). The time of each peak (the center of gravity) was treated as the RT. Relative retention times were calculated for each peak relative to the RT of 13C12-1,2,7,8-TeCDF.
Table 2 shows the retention order of all 136 PCDDs/PCDFs congeners on the two columns. Additionally, the retention order is visually displayed in Figs. 2, 3, 4, 5, 6, 7, 8.
| Homolog | DB-17ms capillary column | VF-17ms capillary column | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Elution order | Congener | RRTa | Elution order | Congener | RRT | Elution order | Congener | RRT | Elution order | Congener | RRT | |
| TeCDD | 1 | 1368 | 0.913 | 12 | 1279 | 1.013 | 1 | 1368 | 0.912 | 12 | 1279 | 1.013 |
| 2 | 1379 | 0.931 | 13 | 2378 | 1.020 | 2 | 1379 | 0.930 | 13 | 2378 | 1.016 | |
| 3 | 1369 | 0.958 | 14 | 1234 | 1.023 | 3 | 1369 | 0.959 | 14 | 1238 | 1.022 | |
| 4 | 1378 | 0.969 | 15 | 1238 | 1.023 | 4 | 1378 | 0.966 | 15 | 1237 | 1.023 | |
| 5 | 1247 | 0.979 | 16 | 1237 | 1.026 | 5 | 1247 | 0.979 | 16 | 1234 | 1.026 | |
| 6 | 1248 | 0.979 | 17 | 1236 | 1.029 | 6 | 1248 | 0.979 | 17 | 1236 | 1.029 | |
| 7 | 1268 | 0.996 | 18 | 1269 | 1.043 | 7 | 1268 | 0.995 | 18 | 1269 | 1.044 | |
| 8 | 1249 | 0.997 | 19 | 1239 | 1.045 | 8 | 1478 | 0.998 | 19 | 1239 | 1.046 | |
| 9 | 1246 | 0.998 | 20 | 1278 | 1.052 | 9 | 1249 | 1.000 | 20 | 1278 | 1.050 | |
| 10 | 1478 | 0.999 | 21 | 1267 | 1.082 | 10 | 1246 | 1.001 | 21 | 1267 | 1.081 | |
| 11 | 1469 | 1.003 | 22 | 1289 | 1.095 | 11 | 1469 | 1.007 | 22 | 1289 | 1.096 | |
| PeCDD | 1 | 12479 | 1.110 | 8 | 12489 | 1.198 | 1 | 12479 | 1.106 | 8 | 12347 | 1.193 |
| 2 | 12468 | 1.111 | 9 | 12347 | 1.198 | 2 | 12468 | 1.106 | 9 | 12489 | 1.193 | |
| 3 | 12368 | 1.148 | 10 | 12467 | 1.199 | 3 | 12368 | 1.141 | 10 | 12467 | 1.194 | |
| 4 | 12469 | 1.152 | 11 | 12346 | 1.212 | 4 | 12469 | 1.150 | 11 | 12378 | 1.204 | |
| 5 | 12478 | 1.160 | 12 | 12378 | 1.213 | 5 | 12478 | 1.152 | 12 | 12346 | 1.211 | |
| 6 | 12379 | 1.167 | 13 | 12367 | 1.237 | 6 | 12379 | 1.160 | 13 | 12367 | 1.229 | |
| 7 | 12369 | 1.190 | 14 | 12389 | 1.253 | 7 | 12369 | 1.186 | 14 | 12389 | 1.247 | |
| HxCDD | 1 | 124679 | 1.298 | 6 | 123469 | 1.359 | 1 | 124679 | 1.291 | 6 | 123469 | 1.355 |
| 2 | 124689 | 1.298 | 7 | 123478 | 1.379 | 2 | 124689 | 1.291 | 7 | 123478 | 1.367 | |
| 3 | 123468 | 1.323 | 8 | 123678 | 1.388 | 3 | 123468 | 1.314 | 8 | 123678 | 1.375 | |
| 4 | 123689 | 1.344 | 9 | 123789 | 1.407 | 4 | 123689 | 1.333 | 9 | 123789 | 1.394 | |
| 5 | 123679 | 1.344 | 10 | 123467 | 1.412 | 5 | 123679 | 1.334 | 10 | 123467 | 1.404 | |
| HpCDD | 1 | 1234679 | 1.502 | 2 | 1234678 | 1.553 | 1 | 1234679 | 1.492 | 2 | 1234678 | 1.538 |
| OCDD | 1.701 | 1.686 | ||||||||||
| TeCDF | 1 | 1368 | 0.879 | 20 | 1678 | 0.989 | 1 | 1368 | 0.879 | 20 | 2467 | 0.991 |
| 2 | 1468 | 0.911 | 21 | 2467 | 0.992 | 2 | 1468 | 0.913 | 21 | 1678 | 0.992 | |
| 3 | 2468 | 0.918 | 22 | 1236 | 0.995 | 3 | 2468 | 0.918 | 22 | 1236 | 0.997 | |
| 4 | 1378 | 0.922 | 23 | 1234 | 0.996 | 4 | 1378 | 0.921 | 23 | 1234 | 0.999 | |
| 5 | 1347 | 0.924 | 24 | 1278 | 1.001 | 5 | 1347 | 0.924 | 24 | 1278 | 1.001 | |
| 6 | 1247 | 0.929 | 25 | 1469 | 1.016 | 6 | 1247 | 0.929 | 25 | 2378 | 1.017 | |
| 7 | 1367 | 0.937 | 26 | 2378 | 1.019 | 7 | 1367 | 0.937 | 26 | 1469 | 1.018 | |
| 8 | 1346 | 0.940 | 27 | 2347 | 1.021 | 8 | 1379 | 0.940 | 27 | 2347 | 1.020 | |
| 9 | 1379 | 0.941 | 28 | 1349 | 1.023 | 9 | 1346 | 0.942 | 28 | 1349 | 1.025 | |
| 10 | 1246 | 0.948 | 29 | 2346 | 1.027 | 10 | 1246 | 0.950 | 29 | 2346 | 1.029 | |
| 11 | 1348 | 0.951 | 30 | 2348 | 1.029 | 11 | 1348 | 0.951 | 30 | 2348 | 1.029 | |
| 12 | 1248 | 0.953 | 31 | 1267 | 1.030 | 12 | 1248 | 0.953 | 31 | 1267 | 1.029 | |
| 13 | 1478 | 0.960 | 32 | 1279 | 1.034 | 13 | 1478 | 0.961 | 32 | 1279 | 1.034 | |
| 14 | 1268 | 0.966 | 33 | 1249 | 1.040 | 14 | 1268 | 0.965 | 33 | 1249 | 1.041 | |
| 15 | 1467 | 0.968 | 34 | 2367 | 1.049 | 15 | 2368 | 0.970 | 34 | 2367 | 1.047 | |
| 16 | 2368 | 0.971 | 35 | 3467 | 1.059 | 16 | 1467 | 0.970 | 35 | 3467 | 1.059 | |
| 17 | 1237 | 0.972 | 36 | 1239 | 1.074 | 17 | 1237 | 0.970 | 36 | 1239 | 1.076 | |
| 18 | 1369 | 0.980 | 37 | 1269 | 1.085 | 18 | 1369 | 0.981 | 37 | 1269 | 1.085 | |
| 19 | 1238 | 0.986 | 38 | 1289 | 1.133 | 19 | 1238 | 0.987 | 38 | 1289 | 1.133 | |
| PeCDF | 1 | 13468 | 1.060 | 15 | 12346 | 1.163 | 1 | 13468 | 1.056 | 15 | 12346 | 1.162 |
| 2 | 12468 | 1.063 | 16 | 12378 | 1.170 | 2 | 12468 | 1.058 | 16 | 12378 | 1.163 | |
| 3 | 13678 | 1.106 | 17 | 12348 | 1.174 | 3 | 13678 | 1.101 | 17 | 12348 | 1.170 | |
| 4 | 12368 | 1.118 | 18 | 12469 | 1.178 | 4 | 12368 | 1.113 | 18 | 12469 | 1.175 | |
| 5 | 12478 | 1.119 | 19 | 12367 | 1.188 | 5 | 12478 | 1.114 | 19 | 12678 | 1.182 | |
| 6 | 13478 | 1.120 | 20 | 12678 | 1.188 | 6 | 13478 | 1.115 | 20 | 12367 | 1.182 | |
| 7 | 13467 | 1.121 | 21 | 12379 | 1.198 | 7 | 13467 | 1.118 | 21 | 12379 | 1.191 | |
| 8 | 12467 | 1.129 | 22 | 23478 | 1.220 | 8 | 12467 | 1.125 | 22 | 23478 | 1.212 | |
| 9 | 13479 | 1.132 | 23 | 12369 | 1.234 | 9 | 13479 | 1.126 | 23 | 23467 | 1.228 | |
| 10 | 14678 | 1.132 | 24 | 23467 | 1.235 | 10 | 14678 | 1.129 | 24 | 12369 | 1.230 | |
| 11 | 12479 | 1.147 | 25 | 12679 | 1.238 | 11 | 12479 | 1.141 | 25 | 12679 | 1.232 | |
| 12 | 12347 | 1.152 | 26 | 12489 | 1.255 | 12 | 12347 | 1.148 | 26 | 12489 | 1.249 | |
| 13 | 23468 | 1.155 | 27 | 12349 | 1.259 | 13 | 23468 | 1.148 | 27 | 12349 | 1.257 | |
| 14 | 13469 | 1.159 | 28 | 12389 | 1.297 | 14 | 13469 | 1.157 | 28 | 12389 | 1.291 | |
| HxCDF | 1 | 123468 | 1.275 | 9 | 123678 | 1.351 | 1 | 123468 | 1.266 | 9 | 123678 | 1.340 |
| 2 | 134678 | 1.283 | 10 | 123479 | 1.364 | 2 | 134678 | 1.274 | 10 | 123479 | 1.354 | |
| 3 | 124678 | 1.284 | 11 | 123679 | 1.386 | 3 | 124678 | 1.275 | 11 | 123679 | 1.374 | |
| 4 | 134679 | 1.301 | 12 | 123469 | 1.386 | 4 | 134679 | 1.292 | 12 | 123469 | 1.380 | |
| 5 | 124679 | 1.319 | 13 | 234678 | 1.397 | 5 | 124679 | 1.309 | 13 | 234678 | 1.385 | |
| 6 | 124689 | 1.338 | 14 | 123689 | 1.400 | 6 | 124689 | 1.327 | 14 | 123689 | 1.389 | |
| 7 | 123478 | 1.343 | 15 | 123789 | 1.458 | 7 | 123478 | 1.332 | 15 | 123789 | 1.446 | |
| 8 | 123467 | 1.346 | 16 | 123489 | 1.472 | 8 | 123467 | 1.338 | 16 | 123489 | 1.463 | |
| HpCDF | 1 | 1234678 | 1.496 | 3 | 1234689 | 1.540 | 1 | 1234678 | 1.482 | 3 | 1234689 | 1.528 |
| 2 | 1234679 | 1.523 | 4 | 1234789 | 1.610 | 2 | 1234679 | 1.511 | 4 | 1234789 | 1.595 | |
| OCDF | 1.747 | 1.729 | ||||||||||






For the 2,3,7,8-congeners, peak resolutions (R) were calculated by using Eq. 1.
| (1) |
where t1: RT of first peak; t2: RT of second peak; w1: peak width at base of the first peak; and w2: peak width at base of the second peak.
TETRA- TO HEXA-CDDsWe constructed chromatograms of the TeCDD congeners (Fig. 2). On DB-17ms, 2,3,7,8-TeCDD was partially separated from the closest congeners (1,2,3,4-/1,2,3,8-TeCDD) (Fig. 2a), with a peak resolution of R=0.6. VF-17ms partially separated 2,3,7,8-TeCDD from the closest congener, 1,2,7,9-TeCDD (Fig. 2b), with R=0.9.
We also constructed chromatograms of the PeCDD congeners (Fig. 3). 1,2,3,7,8-PeCDD was co-eluted with 1,2,3,4,6-PeCDD on DB-17ms (Fig. 3a). In contrast, there was baseline separation of 1,2,3,7,8-PeCDD from the nearest congener, 1,2,3,4,6-PeCDD, on VF-17ms (Fig. 3b), with R=1.9.
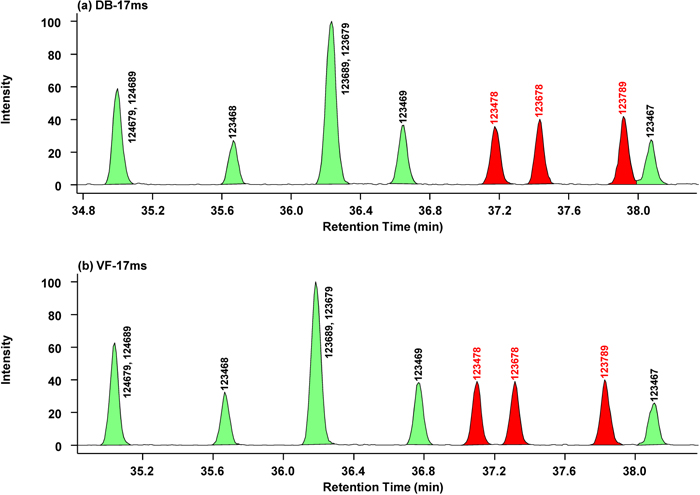
We then created chromatograms of the HxCDD congeners (Fig. 4). The elution order of HxCDDs on DB-17ms and VF-17ms was the same. All 2,3,7,8-chlorine-substituted congeners were separated. DB-17ms gave R values of 1.8 for 1,2,3,4,7,8-HxCDD, 1.8 for 1,2,3,6,7,8-HxCDD, and 1.4 for 1,2,3,7,8,9-HxCDD (Fig. 4a). VF-17ms separated the 2,3,7,8-congeners more clearly, with R=2.0 for 1,2,3,4,7,8-HxCDD, 2.0 for 1,2,3,6,7,8-HxCDD, and 2.2 for 1,2,3,7,8,9-HxCDD (Fig. 4b).
TETRA- TO HEXA-CDFsChromatograms of the TeCDF congeners were constructed (Fig. 5). The elution orders of 2,3,7,8-TeCDF and the nearby congeners differed on the two columns tested. The order on DB-17ms was 1,4,6,9-TeCDF, 2,3,7,8-TeCDF, and 2,3,4,7-TeCDF, and that on VF-17ms was 2,3,7,8-TeCDF, 1,4,6,9-TeCDF, and 2,3,4,7-TeCDF. On DB-17ms, 2,3,7,8-TeCDF was slightly separated from its closest congener, 2,3,4,7-TeCDF (Fig. 5a), with R=0.3. In contrast, 2,3,7,8-TeCDF was co-eluted with 1,4,6,9-TeCDF and 2,3,4,7-TeCDF on VF-17ms (Fig. 5b).
We then created chromatograms of the PeCDF congeners (Fig. 6). The two columns demonstrated good separation of 2,3,4,7,8-PeCDF on DB-17ms (Fig. 6a), with R=3.7, and on VF-17ms (Fig. 6b), with R=3.3. In addition, 1,2,3,7,8-PeCDF was separated on DB-17ms, with a peak resolution of R=1.0, from the closest congener, 1,2,3,4,8-PeCDF. In contrast, 1,2,3,7,8-PeCDF was co-eluted with 1,2,3,4,6-PeCDF on VF-17ms.
Next, we constructed chromatograms of the HxCDF congeners (Fig. 7). DB-17ms separated 1,2,3,6,7,8-HxCDF, 2,3,4,6,7,8-HxCDF, and 1,2,3,7,8,9-HxCDF from the other congeners (R=1.1, 1.0, and 3.2, respectively), and it partially separated 1,2,3,4,7,8-HxCDF (R=0.7). VF-17ms separated 1,2,3,4,7,8-HxCDF, 2,3,4,6,7,8-HxCDF, and 1,2,3,7,8,9-HxCDF from the other congeners (R=1.1, 1.1, and 3.2, respectively); however, 1,2,3,6,7,8-HxCDF was co-eluted with 1,2,3,4,6,7-HxCDF.
HEPTA- AND OCTA-CDDs/CDFsWe also constructed chromatograms of the hepta- and octa-CDD/CDF congeners (Fig. 8a, b). The hepta-CDD/CDF congeners were separated from each other, and the elution orders were the same on the two columns we tested, as on the many columns previously reported (Ryan et al., 1991; Bacher and Ballschmiter, 1992; Matsumura et al., 2003; Fishman et al., 2011). The elution order of the hepta- and octa-CDD congeners was 1,2,3,4,6,7,9-HpCDD and 1,2,3,4,6,7,8-HpCDD, followed by OCDD, whereas that of the hepta- and octa-CDF congeners was 1,2,3,4,6,7,8-HpCDF, 1,2,3,4,6,7,9-HpCDF, 1,2,3,4,6,8,9-HpCDF, and 1,2,3,4,7,8,9-HpCDF, followed by OCDF.
DIFFERENCE BETWEEN DB-17 AND DB-17msThe full assignment of all 136 PCDD/DFs on DB-17, a column analogous to DB-17ms, has been reported by Ryan et al. (1991). DB-17ms is a version of DB-17 designed to MS specifications (Agilent Technologies, 2016). A brief comparison of the two columns is provided here for the reader’s convenience. The 2,3,7,8-congeners that DB-17ms, but not DB-17, can separate are 2,3,7,8-TeCDD, 1,2,3,7,8-PeCDF, and 1,2,3,6,7,8-HxCDF. Conversely, the 2,3,7,8-congeners that DB-17, but not DB-17ms, can separate are 1,2,3,7,8-PeCDD and 2,3,7,8-TeCDF. The remaining 12 congeners can be separated on both columns.
COMBINATION WITH OTHER COLUMNS FOR COMPLETE SEPARATION OF THE SEVENTEEN 2,3,7,8-CONGENERSThe separation of 2,3,7,8-chlorine substituted congeners on the two columns is summarized in Table 3. Six 2,3,7,8-congeners were partially separated or co-eluted on the two columns: 2,3,7,8-TeCDD (DB-17ms and VF-17ms), 1,2,3,7,8-PeCDD (DB-17ms), 2,3,7,8-TeCDF (DB-17ms and VF-17ms), 1,2,3,7,8-PeCDF (VF-17ms), 1,2,3,4,7,8-HxCDF (DB-17ms), and 1,2,3,6,7,8-HxCDF (VF-17ms). Therefore, the two columns tested can be combined with columns capable of separating these six congeners to separate all of the 2,3,7,8-congeners. According to the literature, DB-5ms, VF-5ms, VF-Xms (Fishman et al., 2011), and BPX-DXN (Matsumura et al., 2003) demonstrate good separation of these six congeners. Therefore, their combination (DB-17ms or VF-17ms×DB-5ms, VF-5ms, VF-Xms, or BPX-DXN) should provide complete separation of all seventeen 2,3,7,8-congeners.
| Compound | DB-17ms | VF-17ms |
|---|---|---|
| 2,3,7,8-TeCDD | +− | +− |
| 1,2,3,7,8-PeCDD | −− | ++ |
| 1,2,3,4,7,8-HxCDD | ++ | ++ |
| 1,2,3,6,7,8-HxCDD | ++ | ++ |
| 1,2,3,7,8,9-HxCDD | ++ | ++ |
| 1,2,3,4,6,7,8-HpCDD | ++ | ++ |
| OCDD | ++ | ++ |
| 2,3,7,8-TeCDF | −− | −− |
| 1,2,3,7,8-PeCDF | ++ | −− |
| 2,3,4,7,8-PeCDF | ++ | ++ |
| 1,2,3,4,7,8-HxCDF | +− | ++ |
| 1,2,3,6,7,8-HxCDF | ++ | −− |
| 2,3,4,6,7,8-HxCDF | ++ | ++ |
| 1,2,3,7,8,9-HxCDF | ++ | ++ |
| 1,2,3,4,6,7,8-HpCDF | ++ | ++ |
| 1,2,3,4,7,8,9-HpCDF | ++ | ++ |
| OCDF | ++ | ++ |
++ Peak well separated, R≥1.0; +− peak partially separated, 0.6≤R<1.0; −− peak co-eluted, R<0.6
Here, we presented the complete assignment of all 136 tetra- to octa-CDDs/CDFs separated on two gas chromatography columns, DB-17ms and VF-17ms. Of the seventeen 2,3,7,8-congeners, 2,3,7,8-TeCDD, 1,2,3,7,8-PeCDD, 2,3,7,8-TeCDF, and 1,2,3,4,7,8-HxCDF were partially separated or co-eluted on DB-17ms, and 2,3,7,8-TeCDD, 2,3,7,8-TeCDF, 1,2,3,7,8-PeCDF, and 1,2,3,6,7,8-HxCDF were partially separated or co-eluted on VF-17ms. The other 2,3,7,8-congeners showed good separation on both columns. The concentrations of all seventeen 2,3,7,8-congeners can be obtained by combining the measurement results from DB-17ms or VF-17ms with those from DB-5ms or similar columns.
Currently, the combination of BPX-DXN and RH12ms is the mainstream method for dioxin analysis in Japan (Iwakiri and Enomoto, 2018). Although this combination can separate almost all 2,3,7,8-congeners, the separation of 2,3,4,6,7,8-HxCDF is insufficient (Matsumura et al., 2003). By applying the combination of DB-17ms or VF-17ms with DB-5ms or similar columns, as mentioned above, sufficient separation of all 2,3,7,8-congeners—including 2,3,4,6,7,8-HxCDF—is possible.