Abstract
Objectives: To establish a point-of-care test for coronavirus disease 2019 (COVID-19), we developed a dry loop-mediated isothermal amplification (LAMP) method to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA.
Methods: We carried out reverse transcription (RT)-LAMP using the Loopamp SARS-CoV-2 Detection kit (Eiken Chemical, Tokyo, Japan). The entire mixture, except for the primers, is dried and immobilized inside the tube lid.
Results: To determine the specificity of the kit, 22 viruses associated with respiratory infections, including SARS-CoV-2, were tested. The sensitivity of this assay, determined by either a real-time turbidity assay or colorimetric change of the reaction mixture, as evaluated by the naked eye or under illumination with ultraviolet light, was 10 copies/reaction. No LAMP product was detected in reactions performed with RNA from any pathogens other than SARS-CoV-2. After completing an initial validation analysis, we analyzed 24 nasopharyngeal swab specimens collected from patients suspected to have COVID-19. Of the 24 samples, 19 (79.2%) were determined by real-time RT-PCR analysis as being positive for SARS-CoV-2 RNA. Using the Loopamp SARS-CoV-2 Detection kit, we detected SARS-CoV-2 RNA in 15 (62.5%) of the 24 samples. Thus, the sensitivity, specificity, positive predictive value, and negative predictive values of the Loopamp 2019-CoV-2 detection reagent kit were 78.9%, 100%, 100%, and 55.6%, respectively.
Conclusions: The dry LAMP method for detecting SARS-CoV-2 RNA is fast and easy to use, and its reagents can be stored at 4°C, solving the cold chain problem; thus, it represents a promising tool for COVID-19 diagnosis in developing countries.
Introduction
The outbreak of coronavirus disease 2019 (COVID-19), caused by the novel coronavirus designated as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), started in Wuhan, China, in December 2019.1,2 To date, the outbreak has spread rapidly around the world,1,2 and World Health Organization (WHO) has declared it a pandemic. On 3 March 2022, WHO reported a total of 437,333,859 confirmed COVID-19 cases and 5,960,972 associated deaths. Since SARS CoV-2 was first detected, the original virus has mutated, yielding an impressive array of variants. The accumulation of mutations arising out of subsequent viral replication is a natural phenomenon. WHO defines a variant of interest (VOI) as a variant with genetic changes impacting pathogen transmissibility and disease severity, as well as community transmission in multiple countries. A variant of concern (VOC) shares the characteristics of a VOI but is additionally associated with increased virulence and decreased effectiveness of public health measures. As of November 24, 2021, the five SARS-CoV-2 variants that have been designated as VOC are Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529).
Clinical trials have evaluated several drugs, with remdesivir and systemic corticosteroids showing promise for treating moderate and severe cases of COVID-19, respectively.3 Several COVID-19 vaccines have been authorized; however, COVID-19 vaccine coverage remains insufficient. Antiviral and neutralizing antibody therapies that reduce the risk of COVID-19 progression are needed. New treatments have recently been approved, such as Molnupiravir, nirmatrelvir plus ritonavir (an orally administered antiviral agent targeting SARS-CoV-2),4,5 casirivimab and imdevimab (a combination of two neutralizing monoclonal antibodies [REGEN-COV]),6,7 and sotrovimab (an engineered human monoclonal antibody that neutralizes SARS-CoV-2).8 Several characteristic clinical findings, such as fever and dry cough with typical signs of pneumonia on chest computed tomography,9–11 history of close contact with a patient who has COVID-19 or of visiting a COVID-19-endemic area,12,13 are useful in diagnosing COVID-19. However, many patients have mild symptoms or are asymptomatic, hampering the ability to make an accurate diagnosis on the basis of only clinical features.14–16 Therefore, a rapid diagnostic method is necessary to provide a definitive diagnosis of COVID-19.
Real-time reverse transcription-polymerase chain reaction (RT-PCR) is widely used for the diagnosis of COVID-19.17,18 This method is very useful for testing large numbers of samples at large hospitals, diagnostic companies, and local health facilities. However, a point-of-care (POC) test for COVID-19 is also important for use in the management of patients with suspected cases of COVID-19 in low-resource settings. Additionally, because the expansion of COVID-19 to developing countries is a major public health concern, there is an urgent need to develop rapid diagnostic tests for COVID-19. Performing real-time RT-PCR requires a special thermal cycler with precision optics that monitor fluorescence emission from sample wells. In contrast, the loop-mediated isothermal amplification (LAMP) method can amplify template nucleotides under isothermal conditions with efficiency and specificity as high as those of nested double PCR.19 Owing to its speed, ease of use, and cost-effectiveness, LAMP has been widely used for POC testing for various infectious diseases, including COVID-19.19–22 Furthermore, a dry LAMP reagent mixture, which can be stored at 4°C, is much easier to handle and more heat-stable compared with mixtures using liquid reagents. A dry LAMP reagent kits for the diagnosis of Mycobacterium tuberculosis,23 human African trypanosomiasis,24 malaria,25 and influenza (type A and H1 pdm 2009) have been commercially produced.26 These dry LAMP methods did not require the enzyme or reaction buffer to be aliquoted. Therefore, the dry LAMP method would be very useful for the diagnosis of tropical infectious diseases in regions including Africa,27 which is expected to be the next epicenter of COVID-19.
The aim of this study was to evaluate the performance of the LAMP method using dry reagents for the rapid diagnosis of COVID-19 infection. Although several studies on the use of LAMP to detect SARS-CoV-2 have already been published,28–30 this is the first study to evaluate the performance of dry LAMP reagents for this purpose.
Methods
Viruses and RNA for initial validation analysis
A total of 22 respiratory pathogens were used in our initial validation analysis of the specificity of the study primers. The strain names of these 22 pathogens are listed in Table 1. The Middle East respiratory syndrome coronavirus (MERS-CoV) EMC strain was kindly provided by Dr. Ron A. M. Fouchier, Erasmus Medical Center, Rotterdam, the Netherlands. The SARS-CoV Frankfurt 1 strain was kindly provided by J. Ziebuhr, University of Würzburg, Germany. The clinical isolates of human coronaviruses (HCoV)-HKU1, -OC43, -NL63, and -229E were described by previous studies.31–33 In vitro-transcribed RNA (GenBank accession number MN908947), synthesized using a ScriptMax Thermo T7 Transcription Kit (TOYOBO, Osaka, Japan), was used to determine the detection limit of the Loopamp SARS-CoV-2 Detection kit (Eiken Chemical, Tokyo, Japan).
Table1
Specificity of dry loop-mediated isothermal amplification assay for SARS-CoV-2
| Virus |
Name of isolate |
Amount (copies/reaction) |
| Coronaviruses |
|
SARS-CoV |
Frankfurt-1 |
4.00×107 |
|
MERS-CoV |
EMC |
1.00×105 |
| Human coronaviruses (HCoV) |
|
HCoV-HKU1 |
Tokyo/SGH-15/2014 |
6.00×106 |
|
|
Tokyo/SGH-18/2016 |
1.00×106 |
|
HCoV-OC43 |
VR-1558 |
4.00×108 |
|
|
Tokyo/SGH-36/2014 |
3.00×106 |
|
|
Tokyo/SGH-61/2014 |
1.00×107 |
|
|
Tokyo/SGH-6/2015 |
4.00×106 |
|
|
Tokyo/SGH-65/2016 |
1.00×107 |
|
HCoV-NL63 |
Amsterdam I |
8.00×107 |
|
|
Tokyo/SGH-15/2017 |
1.00×105 |
|
|
Tokyo/SGH-24/2018 |
4.00×104 |
|
HCoV-229E |
VR-740 |
7.00×107 |
|
|
Sendai-H/1121/04 |
1.00×106 |
|
|
Niigata/01/08 |
1.00×105 |
| Influenza virus |
|
A |
A/Texas/50/2012(H3N2) |
1.18×106 |
|
|
A/Narita/1/2009(H1N1) |
2.94×106 |
|
B |
B/Massachusetts/2/2012 |
4.44×106 |
|
|
B/Texas/2/2013 |
1.46×107 |
|
|
B/Brisbane/60/2008 |
2.00×105 |
| Respiratory syncytial virus (RSV) |
|
|
A2 |
1.00×106 |
|
|
B1 |
1.00×106 |
From March 7 to April 30, 2020, when the Wuhan strain of SARS-CoV-2 was prevalent, nasopharyngeal swabs were collected from patients suspected to have COVID-19 at Fujita Health University. Swab samples were collected using a flocked sterile plastic swab applicator and placed directly into 3 mL of BD universal viral transport medium (Becton, Dickinson and Company, Franklin Lakes, NJ, USA). RNA was extracted from the swab samples immediately. This study was approved by the institutional review board of Fujita Health University (No. HM19-493). Written informed consent was obtained from each patient.
RNA extraction
Viral RNA was extracted from 140 μL of BD universal viral transport medium into which a nasopharyngeal swab had been immersed. RNA extraction was performed using the QIAamp Viral RNA mini kit (#52904, QIAGEN, Chatsworth, CA, USA) in accordance with the manufacturer’s instructions. After extraction, RNA was eluted in 60 μL of buffer AVE and stored at –80°C.
Real-time RT-PCR
Real-time RT-PCR assays for detecting SARS-CoV-2 were performed using TaqMan Fast Virus 1-Step Master Mix (#4444436, Thermo Fisher Scientific, Waltham, MA, USA) in accordance with the manufacturer’s instructions. This master mix can perform RT and PCR all in one reaction well. The following primers and probes were used: NIID_2019-nCOV_N_F2, 5'-AAATTTTGGGGACCAGGAAC-3'; NIID_2019-nCOV_N_R2, 5'-TGGCAGCTGTGTAGGTCAAC-3'; and NIID_2019-nCOV_N_P2, 5'-FAM ATGTCGCGCATTGGCATGGA BHQ-3'.34 Single-well denaturation, reverse transcription, and amplification steps were performed on a QuantStudio 1 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA) in the standard mode. Primer and probe concentrations were as follows: NIID_2019-nCOV_N_F2, 500 nM; NIID_2019-nCOV_N_R2, 700 nM; and NIID_2019-nCOV_N_P2, 200 nM. PCR conditions were as follows: reverse transcription at 50°C for 5 min; enzyme activation at 95°C for 20 s; and 45 cycles of denaturation at 95°C for 15 s and primer annealing/extension/fluorescence emission at 60°C for 60 s. The real-time RT-PCR reaction mixture (total volume, 20 μL) contained 5.0 μL of 4× Fast Virus Master Mix, 1.0 μL of primer–probe pre-mix, 5.0 μL of template RNA, and nuclease-free water.
Reverse Transcription-LAMP
Reverse transcription (RT)-LAMP was carried out using the Loopamp SARS-CoV-2 Detection kit (#LMP403, Eiken Chemical, Tokyo, Japan) in accordance with the manufacturer’s instructions. The RT-LAMP method can synthesize cDNA from template RNA and apply LAMP technology to amplify and detect the cDNA with one step in a single tube. Primers were designed to target the N gene and RNA-dependent RNA polymerase (RdRp) gene. Because the entire mixture, except for the primers, is dried and immobilized inside the tube lid, 10 μL of purified RNA and 15 μL of SARS-CoV-2-specific primer sets were added to the bottom of the tube, after which the tube was inverted several times to resuspend the enzyme and buffer. The full reaction mixture was then collected at the bottom of the tube by performing a quick centrifugation. The mixture was subsequently incubated in a real-time turbidimeter (LA-200; Eiken Chemical Tokyo, Japan) for 35 min at 62.5°C. In LAMP, a large amount of DNA is synthesized, yielding a large amount of pyrophosphate ion byproduct. Pyrophosphate ions combine with divalent metallic ions to form an insoluble salt. Adding manganous ion and calcein, a fluorescent metal indicator, to the reaction solution allows the visualization of substantial alterations of the fluorescence during the one-step amplification reaction, which takes 30–60 min.35 For a visual evaluation of the level of fluorescence, the reaction tube was illuminated with an ultraviolet light by using an ultraviolet illumination system (WSE-5300; ATTO, Tokyo, Japan) and was also observed by the naked eye.
Statistics analysis
To determine how the performance of the SARS-CoV-2 dry LAMP assay compared with that of real time RT-PCR assays for detecting SARS-CoV-2, a two-by-two table was created to calculate the sensitivity, specificity, positive predictive value, and negative predictive value of the Loopamp SARS-CoV-2 Detection kit. Analytical performance characteristics with 95% confidence intervals (CI) were calculated for the sensitivity, specificity, positive predictive value, and negative predictive value of the Loopamp SARS-CoV-2 Detection kit compared with the real-time RT-PCR analysis by using GraphPad Prism9 for Windows (GraphPad Software, San Diego, CA, USA).
Results
Specificity and sensitivity of the dry LAMP method for SARS-CoV-2 detection
To determine the specificity of the Loopamp SARS-CoV-2 Detection kit, we tested 22 viruses associated with respiratory infections, including SARS coronavirus, Middle East Respiratory Syndrome (MERS) coronavirus, other human coronaviruses, influenza viruses, and respiratory syncytial viruses. No LAMP product was detected in reactions performed with RNA from these pathogens (Table 1). These results were confirmed by a turbidity assay and agarose gel electrophoresis analysis (data not shown). To determine the sensitivity of the Loopamp SARS-CoV-2 Detection kit, in vitro-transcribed RNAs were serially diluted in 10 mM Tris buffer containing 0.1 mM EDTA, and 50 ng/mL of carrier RNA were used for defining the detection limit. The sensitivity of this assay, as determined by either the turbidity assay or a colorimetric change of the reaction mixtures evaluated by the naked eye, was 10 copies/reaction (Figure 1).
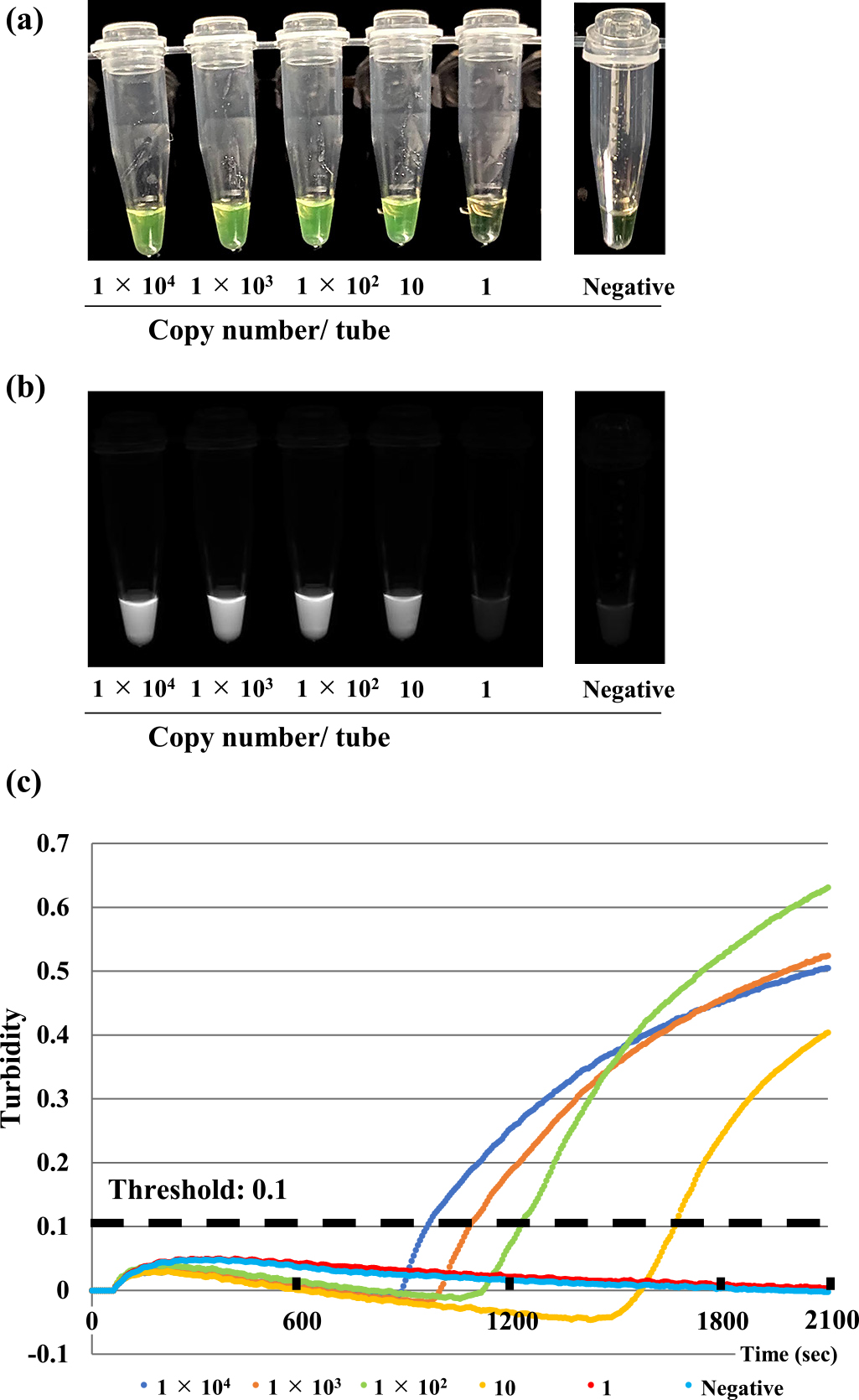
To evaluate the performance of the Loopamp SARS-CoV-2 Detection kit as a POC test, we analyzed 24 nasopharyngeal specimens collected from patients suspected of having COVID-19, including three asymptomatic individuals who came into close contact with COVID-19 cases (Table 2). Of the 24 samples, 19 (78.9%) were determined to be positive for SARS-CoV-2 RNA when assessed by a real-time RT-PCR analysis. When using the Loopamp SARS-CoV-2 Detection kit, SARS-CoV-2 RNA was detected in 15 (62.5%) of the 24 samples. The four false-negative samples contained low copy numbers of viral RNA (2.6, 2.3, 2.2, and 0.5 copies/reaction) and were collected during the convalescent phase of illness (days 7 to 20). Thus, the sensitivity, specificity, positive predictive value, and negative predictive value of the Loopamp SARS-CoV-2 Detection kit were 78.9% (95% CI: 56.7%–91.5%), 100% (95% CI: 56.6%–100%), 100% (95% CI: 79.6%–100%), and 55.6% (95% CI: 26.7%–81.1%), respectively (Figure 2).
Table2
Comparison of real-time RT-PCR and dry loop mediated isothermal amplification assay results of SARS-CoV-2 using clinical specimens
| Case |
Sampling day* |
Real-time RT-PCR
(copies/reaction) |
LAMP |
| 1 |
unknown** |
212732.5 |
+ |
| 2 |
unknown** |
94749.3 |
+ |
| 3 |
7 |
22910.9 |
+ |
| 4 |
5 |
3796.9 |
+ |
| 5 |
11 |
1544.6 |
+ |
| 6 |
10 |
946.9 |
+ |
| 7 |
8 |
413.9 |
+ |
| 8 |
8 |
280.8 |
+ |
| 9 |
9 |
146.0 |
+ |
| 10 |
unknown** |
68.7 |
+ |
| 11 |
12 |
49.8 |
+ |
| 12 |
12 |
48.9 |
+ |
| 13 |
7 |
14.2 |
+ |
| 14 |
8 |
13.2 |
+ |
| 15 |
19 |
4.4 |
+ |
| 16 |
7 |
2.6 |
− |
| 17 |
18 |
2.3 |
− |
| 18 |
10 |
2.2 |
− |
| 19 |
20 |
0.5 |
− |
| 20 |
14 |
0.0 |
− |
| 21 |
8 |
0.0 |
− |
| 22 |
12 |
0.0 |
− |
| 23 |
10 |
0.0 |
− |
| 24 |
12 |
0.0 |
− |
* The day of disease onset was defined as day 1.
** Sampling day could not be determined because the patient was asymptomatic.
Figure 2
Diagnostic performance of the SARS-CoV-2 dry LAMP assay as compared with real time RT-PCR assays for detecting SARS-CoV-2.Abbreviations: +, positive, –, negative, PPV, positive predictive value, NPV, negative predictive value, CI, confidence interval.
|
Real time RT-PCR |
Sensitivity, %
(95% CI) |
Specificity, %
(95% CI) |
PPV, %
(95% CI) |
NPV, %
(95% CI) |
| + |
− |
| LAMP + |
15 |
0 |
78.9
(56.7–91.5) |
100
(56.6–100) |
100
(79.6–100) |
55.6
(26.7–81.1) |
| LAMP – |
4 |
5 |
Discussion
The LAMP assay has several advantages as a POC test method, such as its ease of use, speed, and low cost for amplifying target nucleotides. Accordingly, many investigators have developed SARS-CoV-2 LAMP assays.21,28–30,36 Several improvements to further leverage the advantages of the LAMP method have been also reported, including colorimetric detection using the naked eye29 or a fluorescent detector36 and direct detection of the target sequence without RNA extraction. These improvements shorten and simplify the workflow of the LAMP method29 and enable high-throughput analysis to be conducted. Performing the LAMP method with dry reagents will allow the use of this assay in developing countries because the dry reagents can be stored in refrigerators, which overcomes the requirement of traditional LAMP reagents for strict cold-chain transportation and storage. Therefore, in this study, we investigated the performance of the SARS-CoV-2 dry LAMP method.
No amplification was observed with RNA from other viruses (including SARS coronavirus, MARS coronavirus, and other human coronaviruses associated with respiratory infections) using the SARS-CoV-2 dry LAMP method (Table 1), indicating that this LAMP assay can specifically amplify SARS-CoV-2 RNA. In addition, the detection limit of the kit, based on both the turbidity assay and colorimetric change determined by the naked eye, was 10 copies/reaction, which is almost the same as or slightly higher than that of the previously reported SARS-CoV-2 LAMP assays.36 This initial validation analysis demonstrated that the SARS-CoV-2 dry LAMP method is highly specific and sensitive. Huang et al. showed that SARS-CoV-2 RNA can be amplified by a LAMP assay without RNA extraction,29 which is in line with findings from our previous studies for the detection of herpes simplex virus37 and human herpesvirus-6.38 Therefore, to further improve the LAMP method for use as a POC test, a direct detection method should be developed.
To evaluate the performance of SARS-CoV-2 dry LAMP when analyzing clinical samples, we tested 24 nasopharyngeal swab specimens collected from patients suspected of having COVID-19. The sensitivity and specificity of this assay was sufficient for practical purposes. In 24 nasopharyngeal swab specimens, there are only 5 SARS-CoV-2 RNA-negative samples. While there were four false-negative test results, as shown in Table 2, they were for the specimens with the lowest viral genome copy numbers. Notably, two of the four false-negative samples were collected from patients on days 18 and 20 post-symptom onset to confirm the absence of viral genome prior to hospital discharge. Because all samples with viral loads of more than 4.4 copies/reaction were accurately detected as SARS-CoV-2-positive by both turbidity assay and colorimetric changes, we believe that this SARS-CoV-2 dry LAMP method is sufficiently reliable for clinical use in diagnosing COVID-19.
In summary, this study demonstrated that the SARS-CoV-2 dry LAMP method is a reliable method for use in the rapid diagnosis of COVID-19. The dry LAMP method overcomes the requirement for strict cold-chain transportation and storage of reagents. Therefore, this method is expected to be a useful POC test in developing countries where COVID-19 is spreading.
Acknowledgment
We gratefully acknowledge Eiken Chemical for their contributions to this work.
Notes
Grant Support
This work was supported by the Japan Agency for Medical Research and Development (nos. 19fk0108030j0001, JP19fk0108104, JP19fk0108110, JP19fk0108150, 20fk0108117j0001, and 20fk0108117j0101). This study was also supported by a Health Labour Sciences Research Grant (19HA1003).
Conflicts of Interest
All authors declare no conflict of interest with regard to this work.
References
- 1. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature 2020; 579: 265–269.
- 2. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020; 579: 270–273.
- 3. Bimonte S, Crispo A, Amore A, Celentano E, Cuomo A, Cascella M. Potential antiviral drugs for SARS-Cov-2 treatment: preclinical findings and ongoing clinical research. In Vivo 2020; 34: 1597–1602.
- 4. Hammond J, Leister-Tebbe H, Gardner A, Abreu P, Bao W, Wisemandle W, Baniecki M, Hendrick VM, Damle B, Simón-Campos A, Pypstra R, Rusnak JM. Oral nirmatrelvir for high-risk, nonhospitalized adults with COVID-19. N Engl J Med 2022; 386: 1397–1408.
- 5. Jayk Bernal A, Gomes da Silva MM, Musungaie DB, et al. Molnupiravir for oral treatment of COVID-19 in nonhospitalized patients. N Engl J Med 2022; 386: 509–520.
- 6. Weinreich DM, Sivapalasingam S, Norton T, et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with COVID-19. N Engl J Med 2021; 384: 238–251.
- 7. Weinreich DM, Sivapalasingam S, Norton T, et al. REGEN-COV antibody combination and outcomes in outpatients with COVID-19. N Engl J Med 2021; 385: e81.
- 8. Gupta A, Gonzalez-Rojas Y, Juarez E, et al. Early treatment for COVID-19 with SARS-CoV-2 neutralizing antibody Sotrovimab. N Engl J Med 2021; 385: 1941–1950.
- 9. Liu M, Zeng W, Wen Y, Zheng Y, Lv F, Xiao K. COVID-19 pneumonia: CT findings of 122 patients and differentiation from influenza pneumonia. Eur Radiol 2020; 30: 5463–5469.
- 10. Shi H, Han X, Jiang N, Cao Y, Alwalid O, Gu J, Fan Y, Zheng C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis 2020; 20: 425–434.
- 11. Wang H, Wei R, Rao G, Zhu J, Song B. Characteristic CT findings distinguishing 2019 novel coronavirus disease (COVID-19) from influenza pneumonia. Eur Radiol 2020; 30: 4910–4917.
- 12. Bi Q, Wu Y, Mei S, et al. Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: a retrospective cohort study. Lancet Infect Dis 2020; 20: 911–919.
- 13. Ding J, Tuan WJ, Temte JL. Managing close contacts of COVID-19 confirmed cases in metropolitan areas in China. J Public Health Manag Pract 2020; 26: 345–348.
- 14. Liu YC, Liao CH, Chang CF, Chou CC, Lin YR. A locally transmitted case of SARS-CoV-2 infection in Taiwan. N Engl J Med 2020; 382: 1070–1072.
- 15. Rothe C, Schunk M, Sothmann P, et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med 2020; 382: 970–971.
- 16. Yu P, Zhu J, Zhang Z, Han Y. A familial cluster of infection associated with the 2019 novel coronavirus indicating possible person-to-person transmission during the incubation period. J Infect Dis 2020; 221: 1757–1761.
- 17. Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020; 323: 1843–1844.
- 18. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020; 25: 2000045.
- 19. Notomi T, Mori Y, Tomita N, Kanda H. Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J Microbiol 2015; 53: 1–5.
- 20. Iwamoto T, Sonobe T, Hayashi K. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J Clin Microbiol 2003; 41: 2616–2622.
- 21. Nzelu CO, Caceres AG, Guerrero-Quincho S, Tineo-Villafuerte E, Rodriquez-Delfin L, Mimori T, Uezato H, Katakura K, Gomez EA, Guevara AG, Hashiguchi Y, Kato H. A rapid molecular diagnosis of cutaneous leishmaniasis by colorimetric malachite green-loop-mediated isothermal amplification (LAMP) combined with an FTA card as a direct sampling tool. Acta Trop 2016; 153: 116–119.
- 22. Higashimoto Y, Kawamura Y, Kuboshiki A, Hattori F, Miura H, Nishimura N, Ozaki T, Ihira M, Yoshikawa T. Reliability of direct varicella zoster virus loop-mediated isothermal amplification method for rapid diagnosis of breakthrough varicella. J Clin Virol 2019; 119: 53–58.
- 23. Mitarai S, Okumura M, Toyota E, et al. Evaluation of a simple loop-mediated isothermal amplification test kit for the diagnosis of tuberculosis. Int J Tuberc Lung Dis 2011; 15: 1211–1217, i.
- 24. Njiru ZK, Mikosza AS, Matovu E, Enyaru JC, Ouma JO, Kibona SN, Thompson RC, Ndung’u JM. African trypanosomiasis: sensitive and rapid detection of the sub-genus Trypanozoon by loop-mediated isothermal amplification (LAMP) of parasite DNA. Int J Parasitol 2008; 38: 589–599.
- 25. Polley SD, González IJ, Mohamed D, et al. Clinical evaluation of a loop-mediated amplification kit for diagnosis of imported malaria. J Infect Dis 2013; 208: 637–644.
- 26. Nakauchi M, Yoshikawa T, Nakai H, Sugata K, Yoshikawa A, Asano Y, Ihira M, Tashiro M, Kageyama T. Evaluation of reverse transcription loop-mediated isothermal amplification assays for rapid diagnosis of pandemic influenza A/H1N1 2009 virus. J Med Virol 2011; 83: 10–15.
- 27. Hayashida K, Kajino K, Hachaambwa L, Namangala B, Sugimoto C. Direct blood dry LAMP: a rapid, stable, and easy diagnostic tool for Human African Trypanosomiasis. PLoS Negl Trop Dis 2015; 9: e0003578.
- 28. Dao Thi VL, Herbst K, Boerner K, et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci Transl Med 2020; 12, eabc7075.
- 29. Huang WE, Lim B, Hsu CC, Xiong D, Wu W, Yu Y, Jia H, Wang Y, Zeng Y, Ji M, Chang H, Zhang X, Wang H, Cui Z. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb Biotechnol 2020; 13: 950–961.
- 30. Yu L, Wu S, Hao X, Dong X, Mao L, Pelechano V, Chen WH, Yin X. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin Chem 2020; 66: 975–977.
- 31. Shirato K, Kawase M, Matsuyama S. Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology 2018; 517: 9–15.
- 32. Kazuya S, Nao N, Matsuyama S, Kageyama T. Ultra-rapid real-time RT-PCR method for detecting Middle East Respiratory Syndrome coronavirus using a mobile PCR device, PCR1100. Jpn J Infect Dis 2020; 73: 181–186.
- 33. Shirato K, Kawase M, Watanabe O, Hirokawa C, Matsuyama S, Nishimura H, Taguchi F. Differences in neutralizing antigenicity between laboratory and clinical isolates of HCoV-229E isolated in Japan in 2004–2008 depend on the S1 region sequence of the spike protein. J Gen Virol 2012; 93: 1908–1917.
- 34. Shirato K, Nao N, Katano H, Takayama I, Saito S, Kato F, Katoh H, Sakata M, Nakatsu Y, Mori Y, Kageyama T, Matsuyama S, Takeda M. Development of genetic diagnostic methods for detection for novel coronavirus 2019(nCoV-2019) in Japan. Jpn J Infect Dis 2020; 73: 304–307.
- 35. Tomita N, Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc 2008; 3: 877–882.
- 36. Yan C, Cui J, Huang L, et al. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin Microbiol Infect 2020; 26: 773–779.
- 37. Enomoto Y, Yoshikawa T, Ihira M, Akimoto S, Miyake F, Usui C, Suga S, Suzuki K, Kawana T, Nishiyama Y, Asano Y. Rapid diagnosis of herpes simplex virus infection by a loop-mediated isothermal amplification method. J Clin Microbiol 2005; 43: 951–955.
- 38. Ihira M, Akimoto S, Miyake F, Fujita A, Sugata K, Suga S, Ohashi M, Nishimura N, Ozaki T, Asano Y, Yoshikawa T. Direct detection of human herpesvirus 6 DNA in serum by the loop-mediated isothermal amplification method. J Clin Virol 2007; 39: 22–26.


