2015 年 61 巻 2 号 p. 111-124
2015 年 61 巻 2 号 p. 111-124
Many physiological, biochemical and behavioral processes operate under the circadian rhythm, which is generated by an internal time-keeping mechanism commonly referred to as the biological clock, in almost all organisms from bacteria to mammals. The core circadian oscillator is composed of an autoregulatory transcription-translation feedback loop, in which CLOCK and BMAL1 are positive regulators. A cell has two mechanisms, “cell cycle” and “cell rhythm”, the relationship between which remains controversial. Therefore, the aim of this study was to explore the effect of Clock and Bmal1 on cell cycle, especially on the G1 phase, using vectors with the tetracycline operator-repressor system. The present study revealed that simultaneous induction of Bmal1 and Clock had an influential effect on the cell cycle in SW480/T-REx/Clock/Bmal1 cells, in which both Clock and Bmal1 could be induced by tetracycline. The observation that induction of both Clock and Bmal1 inhibited cell growth and the significant increase of the G1 phase proportion of in SW480/T-REx/Clock/Bmal1 cells indicated that entry from the G1 to S phase was inhibited by the induction of Clock and Bmal1. Furthermore, overexpression of Clock and Bmal1 prevented the cells from entering into the G2/M phase induced by Paclitaxel, and made the cells more resistant to the agent. In conclusion, we found that overexpression of both Clock and Bmal1 suppressed cell growth. In addition, the present study raised the possibility that Clock and Bmal1 may in part play a role in preventing the cells from entering G1 to S phase of cell cycle via suppression of CyclinD1 expression, and thus acquiring resistance to Paclitaxel.
Circadian regulation is a regulation under which many physiological, biochemical and behavioral processes operate. It is generated by an internal time-keeping mechanism commonly known as the biological clock, and exists in almost all organisms from bacteria to mammals1,2). Circadian rhythm is controlled by genetically determined networks of transcription-translation feedback loops involving sets of Clock genes3,4), in which Clock and Bmal1 function as positive regulators, and Period (Per) and Cryptochrome (Cry) act as negative ones5-7). The basic helix-loop-helix-PAS domain proteins CLOCK and BMAL1 form a heterodimer, and then activate transcription of Per, Cry and several other genes besides clock genes through their binding to an E-box (CACGTG) element at each promoter region8,9). In addition, these two molecules are indispensable for maintaining the circadian rhythm feedback loop10,11). After the PER and CRY proteins are translated in the cytoplasm, they form heterocomplexes and translocate into the nucleus and inhibit their own transcription12-14).
In general, living cells are regulated by two different mechanisms, referred to as “circadian rhythm” and “cell cycle”. Both are based on the conceptual device of interlocked autoregulatory loops. Furthermore, both are involved in therapy for patients with certain types of malignancy. “Circadian rhythm” has recently been applied for chronotherapy which contributes to the reduction of side effects and enhancement of chemotherapy. Many studies have reported that the circadian clock could modulate the morbidity and the efficacy of anticancer therapy15,16). These findings led to the idea of chronotherapy as a way of optimizing the efficacy of anticancer drugs for cancer patients by the modification of drug administration at appropriate times of the day. Cell cycle is one of the main targets for anti-cancer drugs. “A large variety of drugs have been developed and analyzed in a cell cycle-dependent manner17-19).
Although some reports have indicated the relationship between circadian rhythm and cell cycle at the G2/M checkpoint20,21), the relationship between circadian rhythm and the G1 checkpoint is still unknown. The aim of this study was to explore the effect of the simultaneous induction of both Bmal1 and Clock on the cell cycle, especially at the G1 checkpoint.
SW480 cells (Human colon cancer cell line) were provided by the Institute of Development, Aging and Cancer, Tohoku University (Sendai, Japan) and cultured in RPMI 1640 (Sigma-Aldrich, St. Louis, MO, USA) containing 10% heat-inactivated fetal bovine serum (FBS) 2 mM glutamine in a 5% CO2 incubator at 37°C. Tetracycline (TET) was purchased from Invitrogen, and its stock solution was prepared in phosphate buffered saline (PBS) at a concentration of 10 mM. Dexamethasone was purchased from Sigma-Aldrich, and its stock solution was prepared in ethanol at a concentration of 2 mM. Blasticidine was purchased from Invitrogen, and its stock solutions were prepared in dimethylsulfoxide (DMSO) at a concentration of 10 mM. Paclitaxel (PTX) was purchased from Sigma, and its stock solution was prepared in DMSO at a concentration of 1 mM.
Cloning of cDNA of Clock and Bmal1, and Plasmid ConstructioncDNA of human Clock and Bmal1 was amplified by PCR using human placental cDNA. The amplified human Clock and Bmal1 cDNA was then cloned into the EcoRI and NotI restriction sites of the pcDNA4/TO vector (Invitrogen, Waltham, MA, USA), respectively. The following primer sequences were used for the cloning of the cDNA: Clock sense primer 5´-TTTAAGCTTTCCTCACAGGAGGCGTGCGG-3´ and antisense primer 5´-TTTAAGCTTTCCTCACAGGAGGCGTGCGG-3´;Bmal1 sense primer 5´-TTTGAATTCATGGCAGACCAGAGAATGG-3´ and antisense primer 5´-TTTCTCGAGTTACAGCGGCCATGGCAAG-3´. The PCR conditions using Pyrobest polymerase (Takara Shuzo, Otsu, Shiga Prefecture, Japan) to amplify the cDNA of Clock were: 10 min at 94°C followed by 35 cycles of 30 s at 94°C, 30 s at 52°C, and 2 min at 72°C, followed by a final cycle at 72°C for 10 min. For amplification of Bmal1 cDNA, the PCR cycling parameters were the same as above, except for the use of LA Taq polymerase (Takara Shuzo) pcDNA4/TO.GFP. Bmal1 and pcDNA4/TO. RFP.Clock were generated by PCR amplification with primers containing in-frame BamHI and EcoRI site, and cloning into identically digested pcDNA4/TO. Bmal1 and pcDNA 4/TO.Clock.
Establishment of Stable Bmal1/ Clock Transfected Cell LinespcDNA6/TR vectors (Invitrogen) were transfected into SW480 cells using Lipofectamine-Plus reagent (Invitrogen) according to the manufacturer’s instructions. SW480-pcDNA6/TR cells were selected by 100 µM of Blasticidine (Invitrogen). pcDNA4/TO.RFP.Clock and pcDNA4/TO.GFP.Bmal1 were transfected into SW480-pcDNA6/TR using Lipofectamine-Plus reagent (Invitrogen) according to the manufacturer’s protocols, and Zeocine (100 µM)-resistant clones were isolated using standard procedures. The pcDNA4/ TO.RFP.Clock and pcDNA4/TO.GFP.Bmal1 co-transfected cells were isolated and examined by fluorescent imaging using an OLYMPUS IX81 microscope with a GFP/Rhodamine filter set (IX2-RFAL and IX2-RFACA, OLYMPUS, Tokyo, Japan) after treatment of the cells with 30 µg/ml TET for 0-48 h. The stable transfected cell line was established after three rounds of subcloning. This cell line was defined as SW480/T-REx/Clock/Bmal1.
Production of Circadian Rhythm by Treatment with DexamethasoneThe method for induction of the circadian rhythm by treatment with dexamethasone was previously described22). SW480 cells were treated with 100 nM dexamethasone for 2 h, and then the medium was replaced with growth medium.
Conventional RT-PCRTotal RNA was extracted using TRIzol (Invitrogen) from cells, and cDNA was synthesized with 2 µg of total RNA and random hexadeoxynucleotide primer (Invitrogen) in 20 µl of a solution containing reverse transcriptase. After synthesis, the cDNA was diluted 1: 4 with distilled water and stored at −20°C until use. Each PCR was performed with cDNA derived from 20 ng of RNA. PCR reactions were carried out in a total volume of 20 µl containing cDNA, dGTP, dATP, dTTP, and dCTP at a concentration of 200 µM, 4 µM of each primer and 0.25 units of ExTaq polymerase (Takara Shuzo). The PCR primer used in this study is listed in Table 1. The PCR cycling parameters were as follows: 10 min at 94°C followed by 35 cycles of 30 s at 94°C, 30 s at 55°C, and 2 min at 72°C, followed by a final extension at 72°C for 10 min. The PCR products were electrophoresed on a 2% agarose gel.
For quantification, cDNA was used as a template in a real-time RT-PCR assay (Bio-Rad Laboratories, Hercules, CA, USA), according to the manufacturer’s protocol. G3PDH served as an internal control and was amplified parallel with every sample. The oligonucleotide primers for the detection of every gene expression and the G3PDH gene are listed in Table 1. The constituents of each PCR (15 µl) were 1 µl of cDNA, 1.0 µl of primer 15 pmol/liter each, 5.5 µl of distilled H2O, and 7.5 µl of iQ SYBR Green Supermix (Bio-Rad Laboratories).
Cell Count AssaySW480 and SW480/T-REx/Clock/Bmal1 cells were plated at a density of 3.0 × 104/25 cm2. When the cells were attached to the bottom of the flask, the medium was changed to growth medium with 30 mg/ml TET. After 48 h, the cells were harvested and counted every 24 h.
Flow CytometrySamples were prepared for flow cytometry. Briefly, cells were harvested, washed in ice-cold PBS (pH 7.4), fixed in ice-cold 70% ethanol, and treated with RNase (500 units/ml; Invitrogen) at 37°C for 20 min. Cellular DNA was stained with 50 µg/ml of propidium iodide (Sigma Chemical Co., St. Louis, MO, USA), and cells were stored at 4°C. Cell cycle analyses were performed using a Becton Dickinson fluorescence-activated cell analyzer (Becton Dickinson, Mansfield, MA, USA). Cells (1 × 104) were analyzed for each point, and quantization of cell cycle distribution was performed using Modfit LT (Verity Software House, Topsham, ME, USA). All experiments were performed when cells reached 50% confluence.
ImmunoblottingCells were lysed with ice-cold RIPA lysis buffer (150 ng NaCl, 5 mM Tris-HCl at pH 7.2, 1% Nonidet P-40, 1% sodium-deoxycorate, 0.05% Sodium dodecyl sulfate) and a protease inhibitor cocktail (Sigma-Aldrich). Protein concentrations were adjusted by the cell number, 2.60 × 106 cells/1 ml RIPA buffer. Proteins were separated in 8% SDS-PAGE and transferred to PVDF membrane (Bio-Rad Laboratories). The transferred proteins were reacted with anti-RFP polyclonal antibody (Medical & Biological Laboratories CO, Nagoya, Japan), anti-GFP N-terminal developed in rabbit affinity isolated antibody (Sigma-Aldrich), and alpha-tublin (Sigma-Aldrich) at 4°C over night, followed by incubation for 30 min at room temperature with horseradish peroxidase- conjugated goat anti-rabbit IgG (Vector Laboratories, Burlingame, CA, USA) or horseradish peroxidase-conjugated goat antimouse IgG (Vector Laboratories). Antibody binding was visualized with the ECL system (GE Healthcare, Little Chalfont, Buckinghamshire, UK).
ImmunoprecipitationThe cells were washed with ice-cold PBS, and the cytosol was lysed with buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 0.5 mM DTT) with proteinase inhibitor cocktail (Sigma-Aldrich). Deposits were lysed with buffer C (20 mM HEPES [pH 7.9], 20% (v/v) glycerol, 0.6 M KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT) with proteinase inhibitor cocktail (Sigma-Aldrich). The supernatants were diluted using distilled water and transferred to a fresh tube, and 2.5 µg of anti-GFP antibody (Sigma-Aldrich) or non-specific rabbit IgG was added, followed by incubation at 4°C for 1.5 h. Then, 30 µl of Protein G Sepharose (GE healthcare) was added and the mixture was incubated at 4°C for 1.5 h. The supernatants were discarded carefully, 1X sodium dodecyl sulfate sample buffer was added, and the samples were boiled for 5 min. They were then separated in 8% SDS-PAGE and transferred to a PVDF membrane (Bio-Rad Laboratories). Immunoblotting was performed with Anti-RFP antibody (Medical & Biological Laboratories CO) or anti-GFP antibody (Sigma-Aldrich) as a primary antibody, and horseradish peroxidase-conjugated anti-rabbit antibody as a secondary antibody. Antibody binding was visualized with the ECL system (GE Healthcare).
MTT AssaysThe cytotoxicity of the anticancer drugs was measured using the MTT colorimetric assay (Sigma Chemical Co.). The cells were seeded in a 96-well plate with growth medium. On the following day, the medium was changed to 180 µl of RPMI 1640 containing 10% FBS and 30 µg/ml of TET. Thereafter, 20 μl of PTX at each concentration (30 nM-30 µM) was added to the wells (the final concentrations were 3 nM-3 µM) and incubations were continued at 37°C for four days, after which 50 µl of MTT (1 mg/ml PBS) was added to each well. After 4 h additional incubation at 37°C, the resulting formazan was dissolved in 90 µl of DMSO. Optical densities were read immediately at 570 nm by using a Micro Plate Reader (Bio-Rad Laboratories).
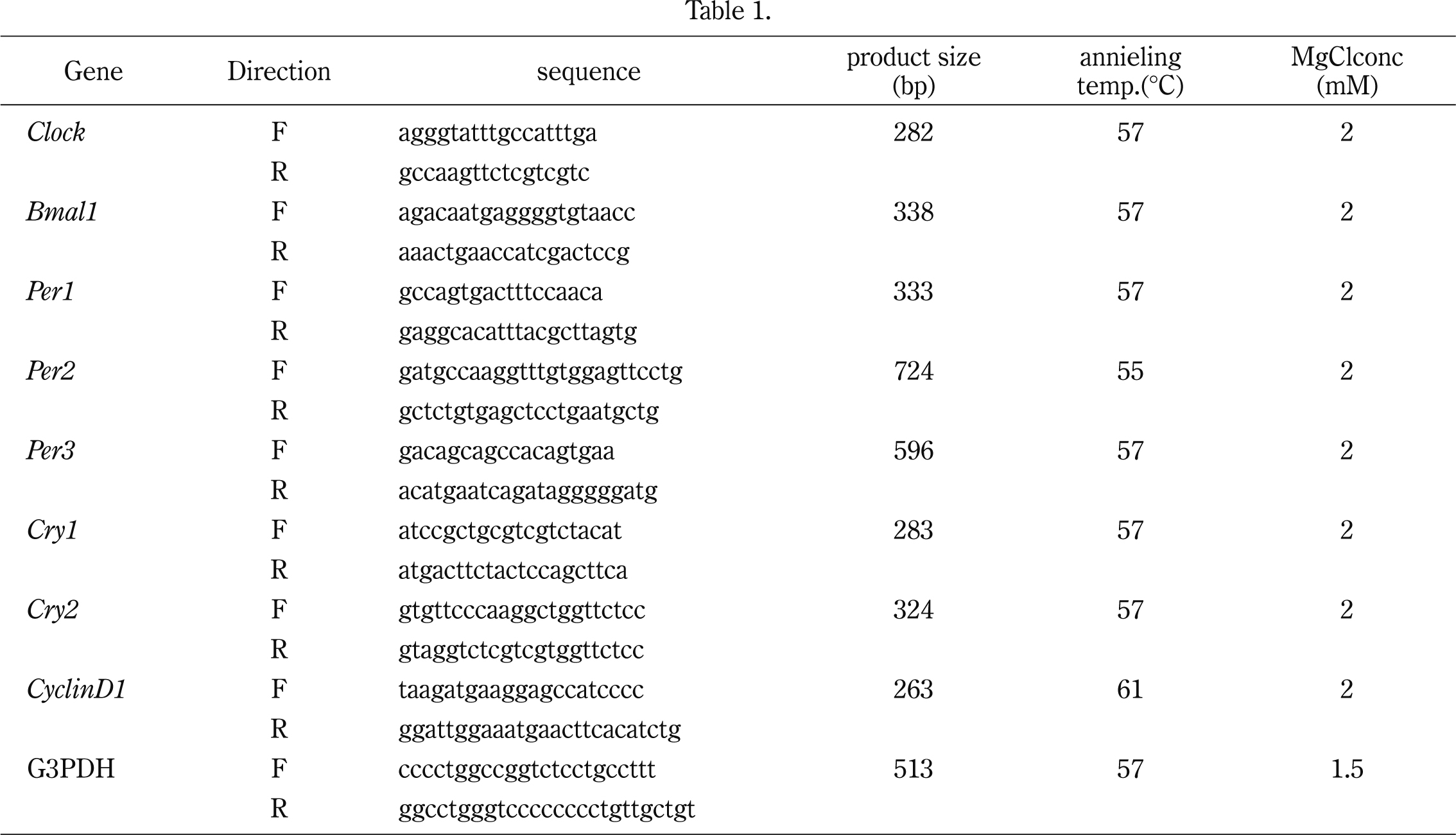
We investigated whether SW480 cells show rhythm under dexamethasone treatment as described in the Materials and Methods section. The mRNA expression of circadian rhythm-related genes including Clock, Bmal1, Per1, Per2, Per3, Cry1, and Cry2 was examined by conventional RT-PCR (Fig. 1A). The mRNA levels of almost all genes were changed depending on time after treatment. We further performed real-time RT-PCR using the same samples (Fig. 1B). The mRNA levels of Bmal1, Per1, and Cry1 were changed periodically, while Clock expression was not changed significantly. Per1 mRNA expression seemed to be inversely correlated with Bmal1 mRNA level.
Circadian Rhythm in SW480 cells
Clock, Bmal1, Per1, Per2, Per3, Cry1, and Cry2 mRNA expressions of the SW480 cells after dexamethasone treatment were determined by conventional PCR (A) and real-time PCR (B). Each bar represents the mean ± SE (bars) of three independent experiments. Statistical differences were calculated by Student’s t-test. Bmal1, Per1, Per3, Cry1, Cry2 had rhythmical mRNA expression, whereas Clock did not (A, B). The expression of Per2 was downregulated in this cell line (B).
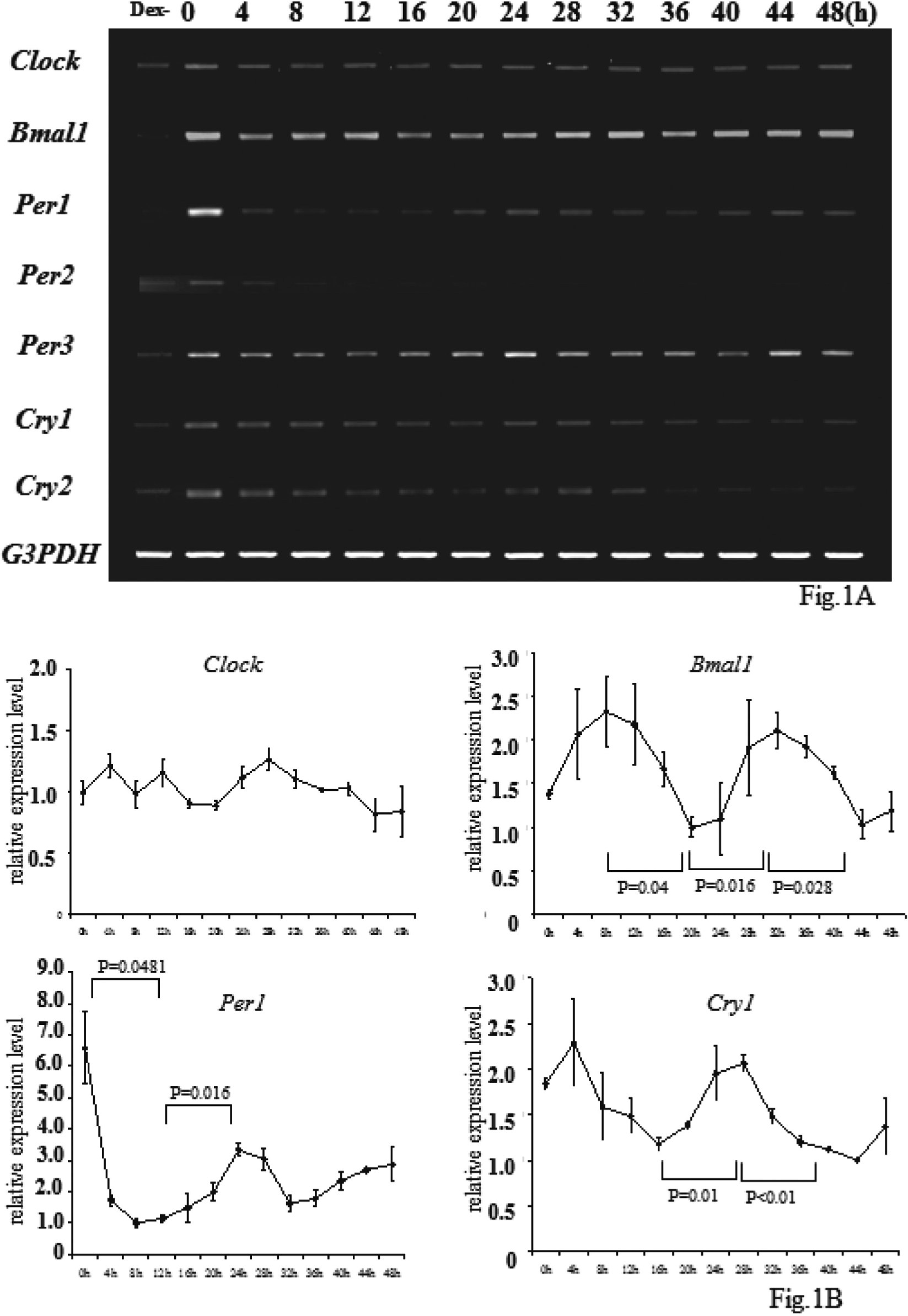
To observe the effects of Clock and Bmal1 on the cell cycle, vectors including Clock or Bmal1, in which TET regulated the target gene expressions, were constructed as described in the Materials and Methods section. The expression of both Clock and Bmal1 at the time after treatment with 30 μg/ml TET in the SW480/T-REx/Clock/Bmal1 cells, was examined by conventional RT-PCR (Fig. 2A), real-time PCR (Fig. 2B), fluorescence microscopy and immunoblotting (Figs. 2C and 2D). The CLOCK/BMAL1 heterodimer activates transcription of Per1 through binding to an E-box element in the Per1 promoter region9);therefore, Per1 expression was used as a positive control in Figs. 2A and 2B. Both Clock and Bmal1 expression levels were increased in a time-dependent manner with an increase of Per1 after TET treatment in SW480/T-REx/Clock/Bmal1 cells. Immunoprecipitation showed a CLOCK and BMAL1-formed heterodimer in our new cell line as already described8,9) (Fig. 2E).
Establishment of Stable Both Clock and Bmal1 Transfected Cell Line
SW480 cells and SW480/T-REx/Clock/Bmal1 cells were plated at a density of 5.0 × 105/60 mm dish. After 24 h, the medium was changed to RPMI1640 supplemented 10% FBS and 30 μ the TET and harvested by TRIzol (Invitrogen) at 0, 3, 6, 12, 18, 24, 36, and 48 h after the medium change. cDNA was synthesized as described above. The expression of Bmal1 or Clock in the cells was observed by conventional RT-PCR (A), real-time PCR (B), fluorescence microscopy and immunoblotting (C). These data show that the mRNA of Clock and Bmal1 was gradually induced, CLOCK and BMAL1 was produced gradually after TET treatment in the SW480/T-REx/Clock/Bmal1 cells (D). Immunoprecipitation showed the CLOCK and BMAL1-formed heterodimer as already described (E).
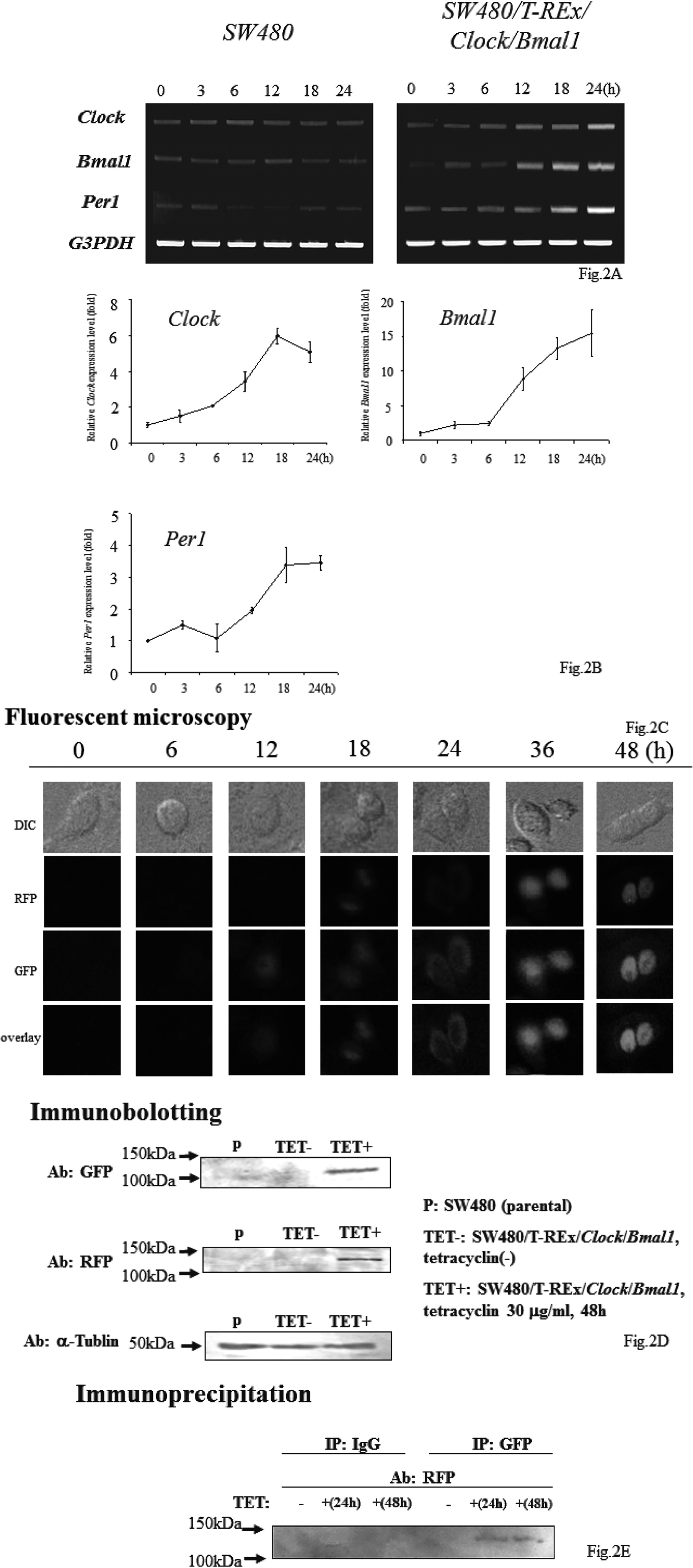
After establishment of the SW480/T-REx/Clock/Bmal1 cells, we performed a cell count assay (Fig. 3). Cell growth was remarkably suppressed by both Clock and Bmal1 induction. At 72 h, the growth ratio of the SW480/T-REx/Clock/Bmal1 cells was significantly less than that of the SW480 cells (5.32 ± 0.82 vs 2.57 ± 0.49, p=0.038).
Overexpression of Both Clock and Bmal1 Suppressed Cell Growth
Each bar represents the mean ± SD (bars) of three independent experiments. Statistical differences were calculated by using Student’s t-test.
To determine how the induction of these two genes suppresses cell growth, we first explored the effect of their induction on the cell cycle by flowcytometry. The proportions of the cells in G1, S and G2/M phase 24 h after TET treatment were 63.59 ± 0.49, 25.90 ±0.81 and 10.51 ± 0.46% in the SW480 cells, and 68.40 ± 0.84%, 18.91 ± 0.63 and 12.69 ± 0.28% in the SW480/T-REx/Bmal1/Clock cells. After 48 h, the G1, S, G2/M proportions were 58.73 ± 1.19, 31.86 ± 1.68% and 9.40 ± 0.70% in the SW480 cells, and 73.15 ± 1.62%, 20.75 ± 1.19 and 6.11 ± 0.48% in the SW480/T-REx/Clock/Bmal1 cells (Fig. 4A). The proportion of G1 phase with and without TET treatment in both cell lines was plotted (Fig. 4B). The proportion of the cells in G1-phase in the SW480/T-REx/Clock/Bmal1 cells (68.40 ± 0.84 % at 24 h, 73.15 ± 1.62% at 48 h) was significantly higher than that in the SW480 cells (63.59 ± 0.49% at 24 h, 58.73 ± 1.19% at 48 h) (p=0.028 at 24 h, p=0.001 at 48 h) (Fig. 4B). To demonstrate increased G1 proportion by Clock and Bmal1 induction more clearly, we performed flow cytometry with cell cycle synchronization by serum starvation (Supplementary Fig. 1). These data indicated that overexpression of both Clock and Bmal1 had an inhibitory effect on entry to the S phase.
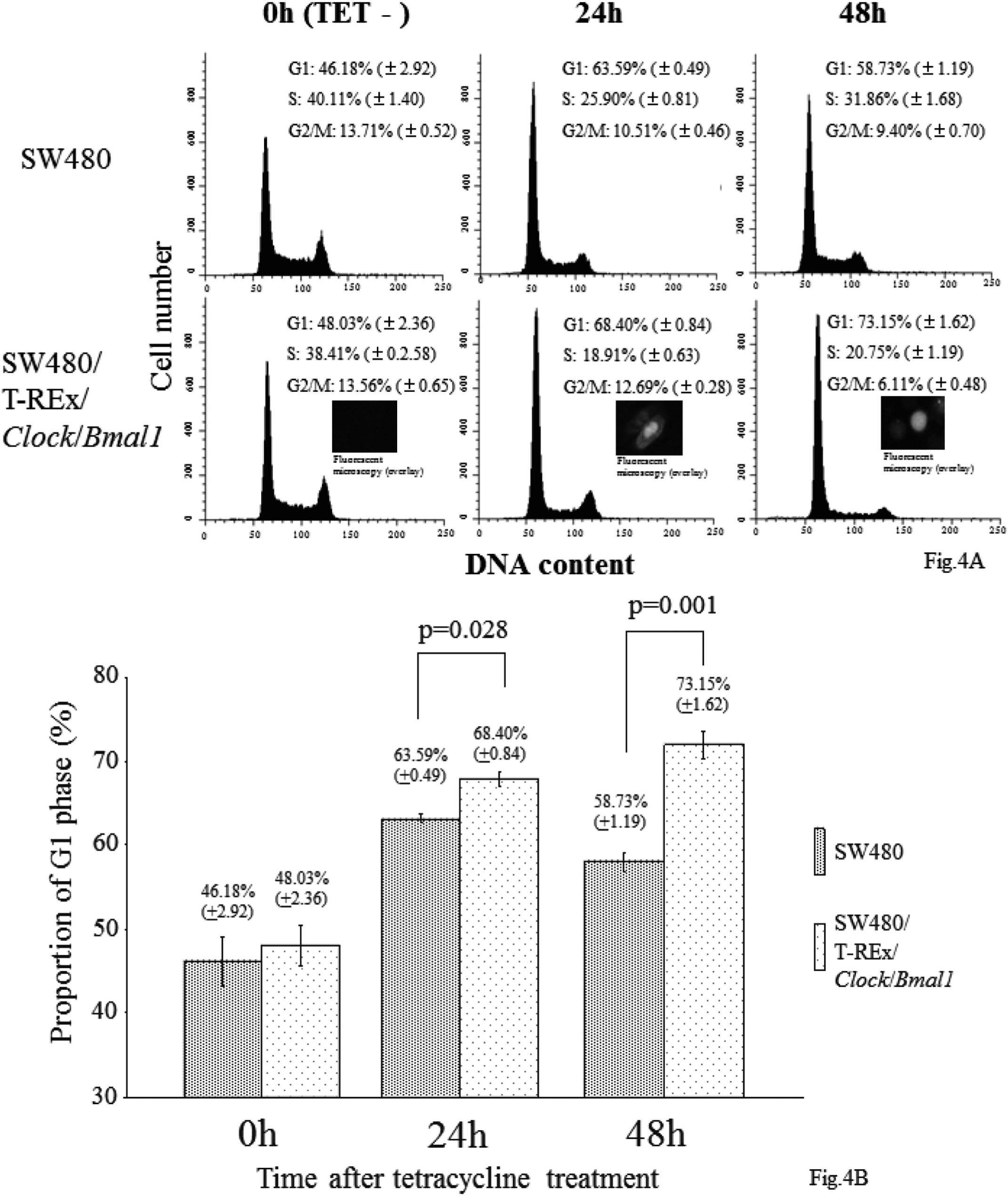
Effect of Both Clock and Bmal1 Induction On Cell Cycle
SW480 cells and SW480/T-REx/Clock/Bmal1 cells were plated at a density of 6.0 × 105/75 cm2. After 48 h, the medium was changed to RPMI 1640 supplemented 30 μg/ml TET and 10% FBS. The samples were harvested and fixed for flow cytometry, as described above, every 24 h. A. Flow cytometric analysis showed a marked increased G1 phase cell proportion by the induction of Clock and Bmal1 in the SW480/T-REx/Clock/Bmal1 cells compared with the parental SW480 cells. Both cell lines were treated with 30 μg/ml TET for 0-48 h. B. The proportions of G1 phase in both cell lines were plotted. Each bar represents the mean ± SD (bars) of three independent experiments. Statistical differences were calculated by using Student’s t-test.
To investigate how overexpression of Clock and Bmal1 inhibits entry to the S phase, we examined CyclinD1 expression level by real-time RT-PCR (Fig. 5). Before TET treatment, the CyclinD1 expression level of the SW480/ T-REx/Clock/Bmal1 cells was the same as that of the SW480 cells (1.0 ± 0.14 vs 1.06 ± 0.13, N.S.), but after TET treatment, the CyclinD1 expression level of the SW480/T-REx/Clock/Bmal1 cells was decreased in comparison with that of the SW480 cells in a time-dependent manner. At 48 h after TET treatment, the CyclinD1 expression level of the SW480/T-REx/Clock/Bmal1 cells was significantly decreased (0.70 ± 0.05 vs 0.38 ± 0.03, p=0.0048). These data suggested that both Clock and Bmal1 overexpression might suppress CyclinD1 expression directly or indirectly, and that entry to the S phase was inhibited due to the decrease of CyclinD1 expression.
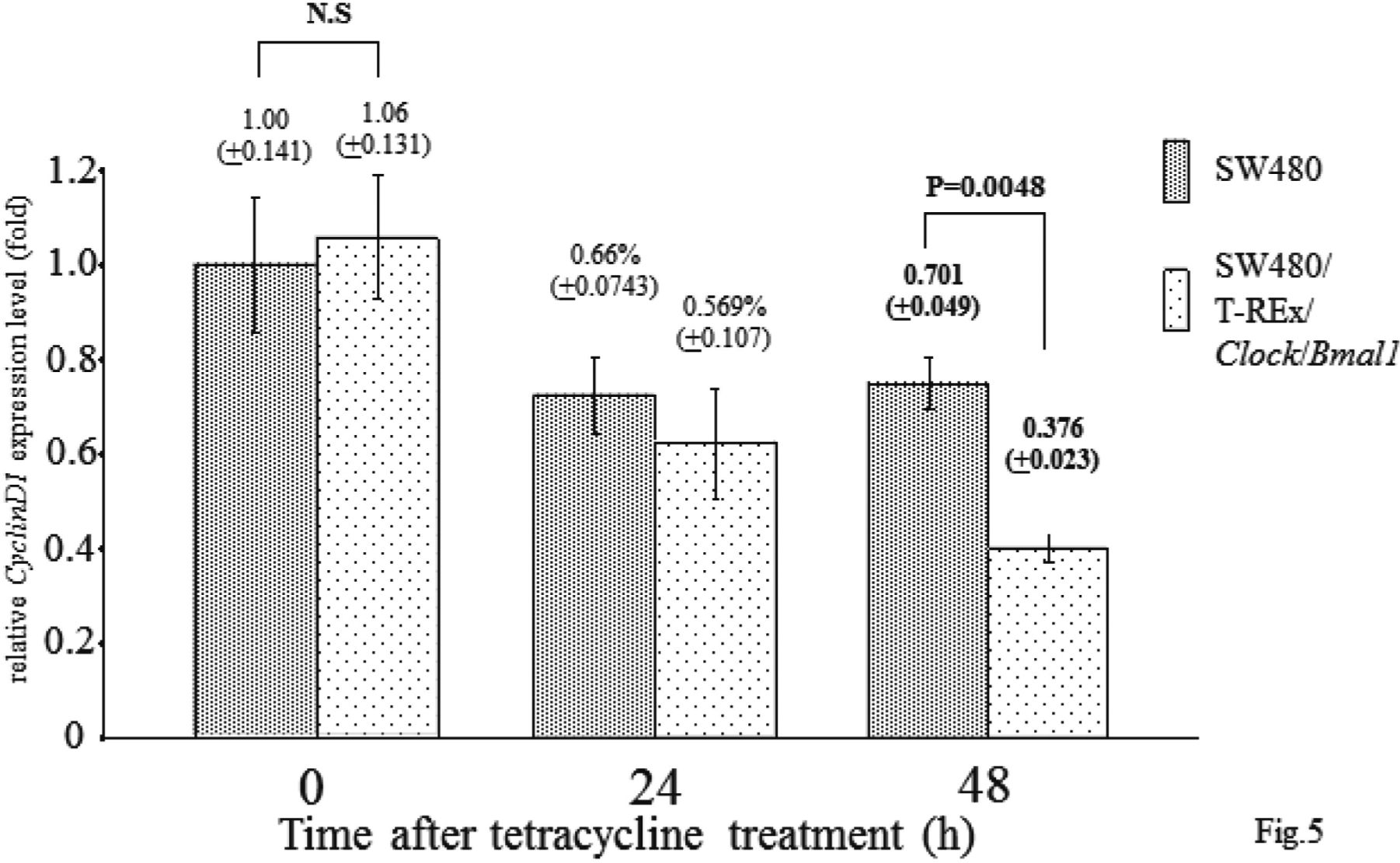
CyclinD1 Expression Level of Clock and Bmal1 Overexpressed Cells
SW480 cells and SW480/T-REx/Clock/Bmal1 cells were treated with 30 μg/ml TET for 0-48 h, and each CyclinD1 expression level was observed by real-time PCR. CyclinD1 was downregulated by Clock and Bmal1 induction. Each bar represents the mean ± SD (bars) of three independent experiments. Statistical differences were calculated by using Student’s t-test.
PTX, a clinically active anticancer agent isolated from the bark of the Pacific yew tree, inhibits mitotic spindle assembly or function, and the progression of mitotic cells to the G1 phase18). If over-expression of both Clock and Bmal1 has an inhibitory effect on entry to the S phase, the effect could compete for the pharmacological mechanism of PTX. We examined whether induction of these two genes altered the sensitivity to PTX. Without TET treatment, PTX inhibited cell cycle progression in the G2/M phase of both the SW480 and SW480/T-REx/Clock/Bmal1 cells equally. However, 24 h following TET treatment, the proportion of the G2/M phase in the SW480/T-REx/Clock/Bmal1 cells was significantly lower than that in the SW480 cells (46.22 ± 1.09% vs 33.66 ± 1.63%, p=0.0127) (Figs. 6A and B). Furthermore, MTT assay showed that the induction of these two genes made the SW480 cells resistant to PTX (Fig. 7). Although TET itself has an effect on the cell cycle to arrest at the G1 phase, the present data showed that simultaneous induction of Clock and Bmal1 changes the chemosensitivity to PTX by inhibiting entry to the G2/M phase in SW480/T-REx/Clock/Bmal1 cells.

Overexpression of Clock and Bmal1 Prevents the Cells from Entry to G2/M Phase by Paclitaxel
A. SW480 cells and SW480/T-REx/Clock/Bmal1 cells were plated at a density of 6.0 × 105/75 cm2. (A) Cells were not pretreated with TET, but treated with PTX for 24 h, (B) Cells were not pretreated with TET but treated with 100 nM PTX and 30 μg/ml TET for 24 h, (C) cells were pretreated with 30 μg/ml TET for 24 h and treated with 100 nM PTX and 30 μg/ml TET for 48 h. The samples were harvested and fixed for flow cytometry described above. A. Flow cytometric analysis showed cell cycle arrest by 100 nM PTX in the SW480/T-REx/Clock/Bmal1 cells and the parental SW480 cells. B. The proportion of the G2/M phase was plotted in a graph. Each bar represents the mean ±SD (bars) of three independent experiments. Statistical differences were calculated by using Student’s t-test.
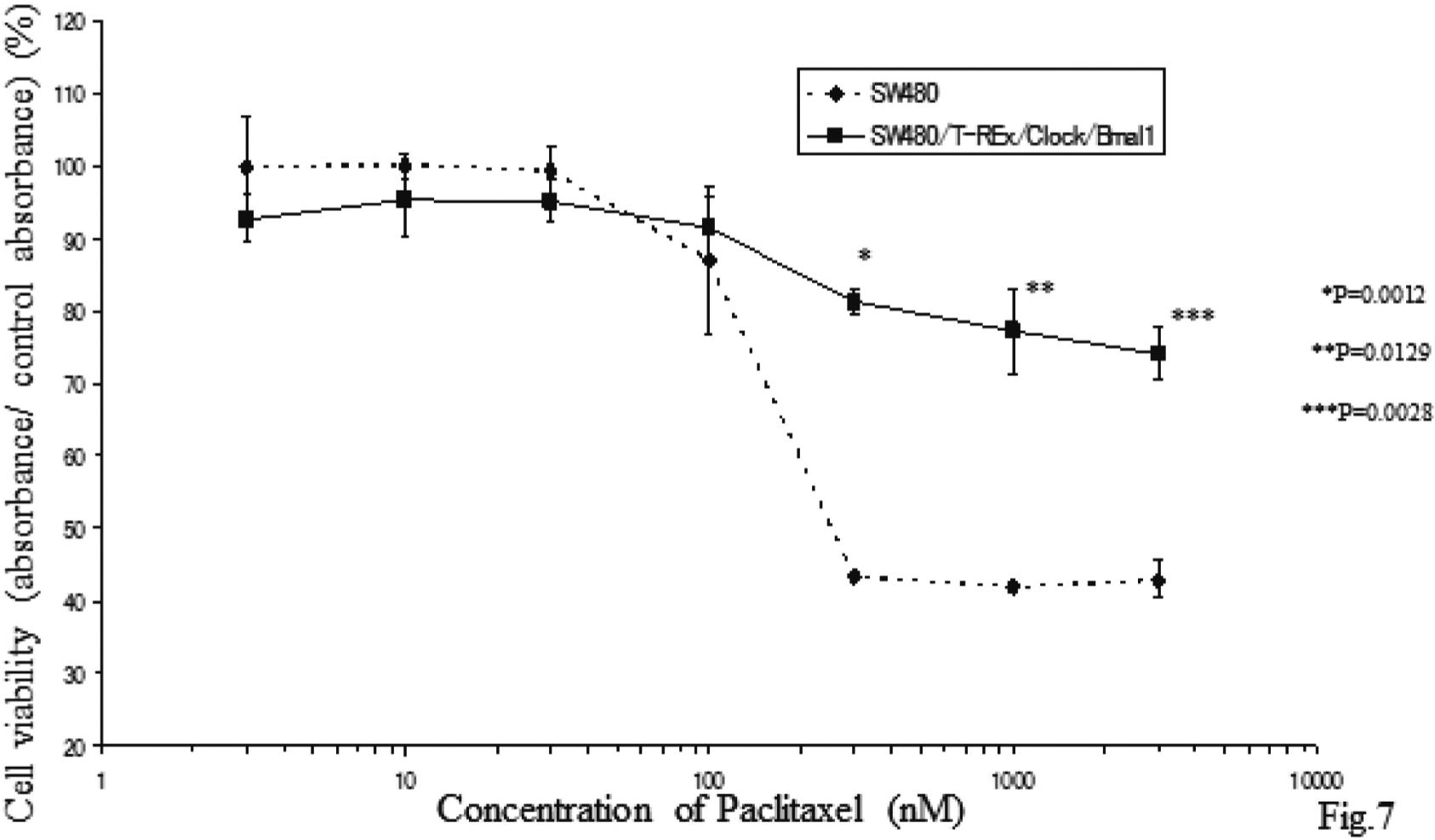
Overexpression of both Clock and Bmal1 Makes SW480 More Resistant to Anticarcinoma Agent Paclitaxel
Each bar represents the mean ± SD (bars) of three independent experiments. Statistical differences were calculated by using Student’s t-test.
We confirmed that Clock and Bmal1 form a heterodimer and promote Per1 expression using SW480/T-REx/Clock/Bmal1 cells. It is important to note “both Clock and Bmal1” not “either Clock or Bmal1”, because the two proteins form a heterodimer, and bind to an E-box element at the promoter region of the target genes9). We showed that CLOCK and BMAL1 made a complex by immunoprecipitation. Kwon et al. reported that singly expressed CLOCK proteins were located in the cytoplasm but when CLOCK and BMAL1 were co-expressed they were found in the nucleus23; thus, Fig. 2C indicates that CLOCK and BMAL1 were dimerized in our cell line. One of the advantages of using the T-RExTM system is the gradual induction of the target genes in a time-dependent manner; we can observe the change that the genes cause more precisely by chasing the time course.
The major findings of the present study were as follows; i) overexpression of both Clock and Bmal1 simultaneously inhibited cell growth, ii) the mechanism of slow cell growth was an inhibitory effect of these two genes on entry from the G1 phase to the S phase at cell cycle level, iii) in the Clock and Bmal1 overexpressed cells, CyclinD1 was downregulated, which may be a cause of increased G1 proportion, iv) overexpression of both Clock and Bmal1 made cells more resistant to PTX.
We demonstrated that the simultaneous overexpression of both Clock and Bmal1 inhibited cell growth possibly by inhibiting entry from the G1 phase to the S phase at the cell cycle level. To the best of our knowledge, this is the first report to indicate that Clock and Bmal1 are involved in cell growth. Although Gorvacheva et al. have already produced Clock mutant/Bmal1 knockout mice, they did not report any growth abnormalities of the mice16). Matsuo et al. reported that liver regeneration of Cry1−/− and Cry2−/− mice is slower than that of wild type mice20). In comparison with these reports, our data is not necessarily contradictable, because in Cry1−/− and Cry2−/− mice, Clock and Bmal1 expression levels should be relatively higher than those in wild type mice. In addition, it has been already reported that circadian clock genes Per1 and Per2 are involved in the G2/M or ATM checkpoints of the cell cycle20,21), but there is yet to be report showing that the circadian gene is directly involved in the G1 checkpoint.
Gery et al. showed that PER1 interacted with Chk2, a key kinase of the ATM checkpoint pathway, and the expression levels of Wee1 and CyclinB1, key molecules for the entry from the G2 phase to the M phase, were downregulated by overexpression of Per121). Matsuo et al. reported that there were three E-box elements in the Wee1 gene 5´-up stream region, and CLOCK and BMAl1 together produced a major increase in transcriptional activity of Wee120). These reports were referred to G2/M checkpoint molecules. The G1 checkpoint is thought to be the most major checkpoint of the cell cycle, and CyclinD is the first response gene that connects growth signal and the start of the cell cycle. The expression of CyclinD1 has reportedly been involved in the circadian system in vitro and in animal models24-27); however, these reports simply showed that expression of CyclinD1 had a circadian rhythm, and did not show that it detailed control mechanisms. Our data suggested that CyclinD1 might be suppressed by the Clock-Bmal1 heterodimer or by other genes activated by the one.
Of importance is the fact that the characteristics of the SW480/T-REx/Clock/Bmal1 cells in the present study that should be mentioned are “gradual induction” and “not so much overexpressed Clock and Bmal1”. In this cell line, Clock and Bmal1 were gradually induced over a period of 24 h, and peak expression levels were about seven times (Clock) and 15 times (Bmal1) of the unsynchronized SW480 expression levels. Therefore, it was not so unnatural that Clock and Bmal1 expression levels were about 10 times those of unsynchronized SW480, in comparison that Bmal1 expression level of synchronized SW480 changed between 1 to 2.5 times (Fig. 1B). Several reports suggest that mouse circadian rhythm, CLOCK and BMAL1 abundance are relatively higher at night25,28,29). The results of the present study may indicate possibility that the administration of PTX at night is not as effective as its daytime administration. To confirm this hypothesis, further studies are required.
In conclusion, our present data raised the possibility that Clock and Bmal1 may have an effect on the G1 checkpoint of the cell cycle either directly or indirectly, and may change the sensitivity to PTX l of the human colon carcinoma cell SW480.
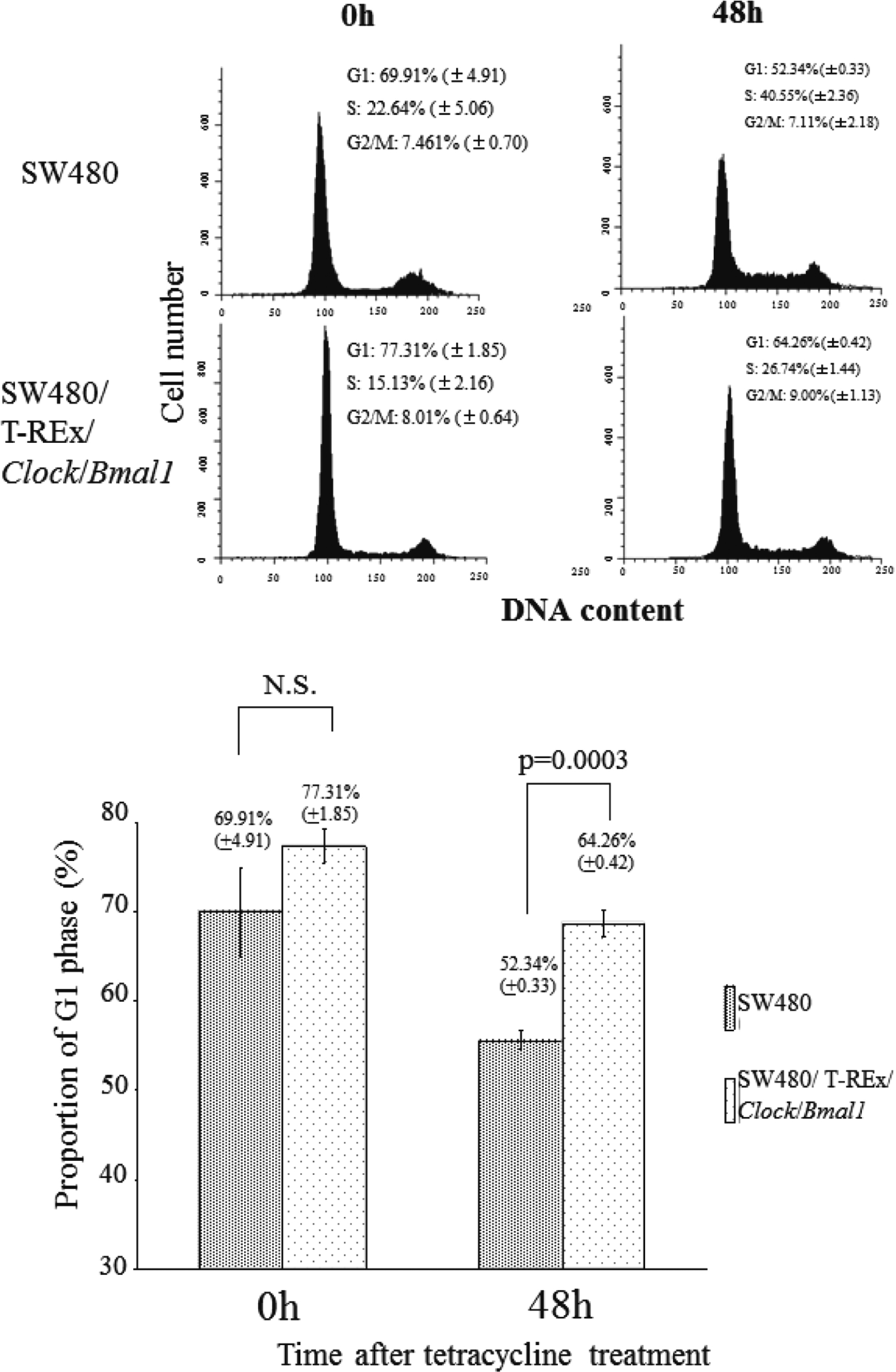
SW480 cells and SW480/T-REx/Clock/Bmal1 cells were plated at a density of 1.0 × 106/75 cm2. After 48 h, the medium was changed to RPMI 1640 medium supplemented 0.2% FBS and 30 μg/ml TET. Then, 48 h after the medium change, the medium changed to a growth medium-supplemented 30 μg/ml TET. The samples were harvested and fixed for flow cytometry every 24 h as described above. Each bar represents the mean ± SD (bars) of three independent experiments. The proportion of G1 phase was plotted in a graph. Statistical differences were calculated by using Student’s t-test. These data strongly suggest that overexpression of both Clock and Bmal1 had an inhibitory effect on entry to the S phase.
none.
The authors would like to thank Prof. Mitsuru Munakata and Dr. Miwako Honma at Fukushima Medical University for their valuable advice in improving this manuscript.
Period1 (Per1), Period2 (Per2), Period3 (Per3), Cryptochrome1 (Cry1), Cryptochrome2 (Cry2), Bmal1 (ARNT-LILE PROTEIN 1, BRAIN AND MUSCLE), GFP (Green Fluorescent Protein), RFP (Red Fluorescent Protein)