2013 Volume 88 Issue 1 Pages 3-7
2013 Volume 88 Issue 1 Pages 3-7
RNA interference is now a well-recognized post-transcriptional mechanism for regulation of gene expression in both animals and plants. In this process, microRNAs (miRNAs) direct silencing complexes to complementary RNA sequences, leading to either degradation or repression of translation. Plants also contain another type of small RNA, small interfering RNAs (siRNAs), that play a role in gene silencing by directing cytosine methylation activities of complementary DNA sequences and thus, differ from miRNAs. This nuclear regulation system is referred to as RNA-directed DNA methylation (RdDM). In plant genomes, transposable elements were initially thought to be regulated by DNA methylation alone. However, several recent reports have revealed that siRNAs and RdDM also play crucial roles in silencing of transposons and endogenous repeats. It is also becoming apparent that transposons are subjected to different levels of regulation in response to developmental and environmental cues. Transposons are tightly regulated in germ cells to protect the host genome from transgenerational mutagenic activity. In plants, transposons are also activated by biotic and abiotic stress. The regulation of transposons in these different situations has been associated with both the DNA methylation and siRNA-mediated regulation systems, suggesting that plants likely evolved “multi-lock” systems for transposon regulation to ensure tight control during the developmental phase and environmental changes.
Plant genomes include many transposon copies. Although many transposons are inactivated by mutations in their DNA sequence, plants face the challenge of keeping the remaining copies transcriptionally silent. Transcriptional silencing is generally achieved by epigenetic modification, particularly DNA methylation (Ichiyanagi, 2013). However, it is not so simple because changes in epigenetic regulation also occur during environmental responses, developmental phase changes, and meiosis that can alter the epigenetic regulation. Recent research has identified additional factors that are essential for the regulation of transposon activity, including complementary small interfering RNAs (siRNAs) and both RNA- and DNA-silencing complexes. This review provides an overview of transposon regulation during developmental and environmental changes and describes the role of siRNAs in ensuring tight control of transposons during these events.
Plant siRNAs are non-coding RNA molecules that are generally 21–24 nucleotides in length and play a role in RNA interference (RNAi) similar to microRNAs (miRNAs; Bartel, 2004). However, unlike miRNAs that are encoded in the genome by precursor hairpin sequences, siRNAs are generated from the cleavage of perfectly complementary double-strand RNA sequences by Dicer-family proteins. In plants, RNAi is thought to have evolved from an anti-viral genome protection mechanism. Higher plants gained specific DNA-dependent RNA polymerases (Kanno et al., 2005; Mosher and Melnyk, 2010; Onodera et al., 2005; Pontier et al., 2005). RNA polymerase IV and V (Pol IV and Pol V) are plant-specific homologs of RNA polymerase II. Pol IV and Pol V produce initial RNA transcripts for RNA silencing and siRNA-induced methylation, respectively. The non-coding RNAs can be transcribed by Pol IV at target loci that include high-copy repeats or transposons. Pol IV is required for the production of > 90% of all siRNAs in Arabidopsis (Matzke and Birchler, 2005; Ream et al., 2009; Zhang et al., 2007). Following transcription by Pol IV, the derived single-stranded RNAs are subsequently converted to double-stranded RNAs by RNA-dependent RNA polymerase 2 (RDR2), which are then processed by DICER-LIKE 3 (DCL3) to generate 24- to 26-nt siRNAs (Mosher et al., 2008; Pontier et al., 2005; Zhang et al., 2007). The RNA-induced silencing complex (RISC) containing ARGONAUTE 4 (AGO4) binds a subset of these siRNAs. The complex scans the nucleus for matching transcripts by base pairing with complementary sequences (Qi et al., 2006). After being loaded with an siRNA, AGO4 interacts with Pol V to recruit the DNA methyltransferase, DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2; Cao and Jacobsen, 2002; Matzke et al., 2004; Matzke and Birchler, 2005), leading to RNA-directed de novo DNA methylation (RdDM) and histone modification of the target. Transposon-derived siRNAs may cause DNA methylation of nearby genes via RdDM and alter histone modification of the locus, leading to heterochromatinization and gene expression regulation.
A major regulator of transposon activation is methylation of DNA sequences. In plants, cytosine methylation in all sequence contexts is controlled by specific DNA methyltransferases. Indeed, loss-of-function mutants of DNA methyltransferases, such as METHYLTRANSFERASE 1 (MET1), release activity of some transposons (Kato et al., 2003; Ronemus et al., 1996). Transposons are also activated in a mutant of SWI2/SNF2-like chromatin remodeling factor, DECREASE IN DNA METHYLATION 1 (DDM1), which maintains CG/non-CG methylation (Hirochika et al., 2000; Miura et al., 2001; Tsukahara et al., 2009). Most transposons are silenced by methylation of their promoters; however, once a transposon is activated by a biotic or an abiotic impact, such as environmental stress, plants use a second-lock system to protect their own genome from the mobile elements. The second regulator is siRNA-mediated de novo DNA methylation and histone modification. The following two reports provided insight into the regulation of transposon mobility in Arabidopsis via siRNA.
Transcription of the Copia93 retrotransposon named “Evade” (EVD) was suppressed by CG methylation, although transpositional activity was independently regulated through a post-transcriptional mechanism involving siRNA and methylation of histone H3 lysine 9 (Mirouze et al., 2009). In met1 mutants, EVD’s transcriptional activity was released, but EVD did not transpose. However, when a met1 mutant was combined with a mutant deficient in siRNA biogenesis, a burst of EVD transposition was observed. This indicates that epigenetic regulation of retrotransposition is controlled by selective machinery involving siRNAs.
When another Copia-type retrotransposon, ONSEN, was activated by heat stress, it was shown to synthesize extrachromosomal DNA that had the potential to transpose (Ito et al., 2011). An increase in transcription level in a mutant deficient in siRNA biogenesis indicated that siRNA regulates ONSEN expression. ONSEN transposition was observed in the progeny of a mutant deficient in siRNA biogenesis but not in wild-type plants, suggesting that the siRNA engaged not only in transcriptional but also in transpositional regulation of ONSEN.
In animals, epigenetic reprogramming, including demethylation of DNA and remodeling of histone modifications, occurs in germ cells and early embryos on a genome-wide scale (Saito, 2013). Suppression of transposon activity in gametes is an important defense to ensure host genome stability by preventing transmission of mutagenic activity to the next generation. To maintain transgenerational genome integrity, plants show meiotic inheritance of gene silencing through the stable inheritance of DNA methylation. However, large-scale reprogramming also occurs in non-germ line reproductive cells.
In Arabidopsis, female gametogenesis produces one egg cell, one central cell (two nuclei), and several other accessory cells. During fertilization, the egg cell fuses with one of the two sperm cells from the male gametophyte to form an embryo, and the central cell fuses with the other sperm cell to form the triploid endosperm. DEMETER (DME) is a helix-hairpin DNA glycosylase and removes methylated cytosines causing global hypomethylation in the endosperm (Gehring et al., 2009). Active demethylation by DME reactivates transposon expression, which introduces transposon transcripts into the RNAi pathway, producing additional siRNAs that can guide DNA methylation (Ibarra et al., 2012). The expression of siRNAs in the development of the endosperm specifically originates from maternal chromosomes, indicating genome imprinting. The siRNAs produced in the endosperm have been reported to move into the egg cell to guide DNA methylation, seemingly to reinforce silencing in the germ cells (Hsieh et al., 2009). This mechanism likely functions in suppressing transposon activity in the embryo; however, “mild” reactivation of transposons in the endosperm may not be deleterious because the endosperm is not inherited by the next generation.
Epigenetic reprogramming also plays an important role in paternal genome reprogramming in Arabidopsis. The male gametophyte contains two sperm cells and a vegetative nucleus. The egg cell and central cell are fertilized by one sperm cell each, resulting in double fertilization. Many genes involved in siRNA biogenesis and silencing are either not expressed or expressed at low levels in pollen. The exception is DDM1, which is exclusively localized in sperm cells but not in the vegetative nucleus of mature pollen (Slotkin et al., 2009). In wild-type vegetative nuclei, the downregulation of DDM1 correlates with DNA demethylation and transposon reactivation. Similar to the endosperm, transposon activation in pollen vegetative nuclei does not impair fitness of the next generation because the vegetative nucleus does not contribute DNA to the fertilized embryo or endosperm. It has been postulated that the benefits of hypomethylation of the vegetative cell occur when small RNAs are transported from the pollen grain cytoplasm into the sperm cells, resulting in siRNA accumulation and de novo remethylation via RdDM to reinforce silencing of transposons in the gametes (Olmedo-Monfil et al., 2010). Calarco et al. (2012) sequenced the methylome of the sperm cell, the vegetative cell, and the postmeiotic microspore and found that, unlike in mammals, the plant germline retains CG and CHG DNA methylation. However, CHH methylation is lost from retrotransposons in microspores and sperm cells and is restored by de novo DNA methyltransferase guided by RdDM machinery, both in the vegetative nucleus and in the embryo after fertilization. These results demonstrated that genome reprogramming in pollen contributes to epigenetic inheritance and transposon silencing guided by small RNAs.
Environmental stress alters chromatin structure that is associated with epigenetic regulation, including histone modification and DNA methylation. Although most transposons are silenced by epigenetic regulation, environmental changes may induce transposon activation (Grandbastien, 2004). Stress can induce changes in gene expression through DNA hypomethylation. For example, cold stress-induced hypomethylation triggers transposition of the Tam-3 transposon in Antirrhinum majus (Hashida et al., 2006). siRNAs and DNA methylation are known to be associated with the Tnt1 transposon in Solanaceae (Andika et al., 2006). Tnt1 can be activated by biotic stress, such as infection and wounding, and also by abiotic stress (Grandbastien et al., 2005). However, the relationship between the activation of Tnt1 and siRNAs under stress is still poorly understood.
More direct evidence of siRNA-mediated regulation has come from research on ONSEN transposition. ONSEN activated by heat stress was transposed to the next generation in an siRNA biogenesis mutant (Ito et al., 2011). ONSEN long terminal repeats (LTRs) contain a common heat-response motif and are responsible for heat-stress induced activation. Under stress, ONSEN is expressed, inducing siRNA-mediated RdDM to resilience the complement target copies (Ito et al., 2011; Matsunaga et al., 2012). In an siRNA biogenesis deficient mutant, ONSEN was activated, and transgenerational transposition was observed, indicating that siRNA is an important regulator of stress-induced transposon activation.
Recently, McCue et al. (2012) reported that transposon-derived siRNAs play a role in the regulation of gene expression and stress response in trans in Arabidopsis. siRNA854, produced from an Athila-family retrotransposon, regulates UBP1b mRNA, which encodes an RNA-binding protein involved in stress granule formation. The regulation occurs on the post-transcriptional and -translational levels when Athila retrotransposons are epigenetically activated, resulting in a phenocopy of the stress-sensitive ubp1b mutant phenotype. Athila is likely to encode siRNA854 to inhibit UBP1b protein formation and interfere with the function of translational repression (McCue et al., 2012).
Plants as a host genome of transposons have evolved several secure systems to protect their genome from ectopic transposition of mobile elements. The first lock is transcriptional regulation by epigenetic modifications that cause repressive heterochromatinization. Most transposons are silenced by this regulatory mechanism. However, the first lock is transiently released by developmental or environmental changes. When a transposon evades transcriptional regulation, plants actuate a second lock, an siRNA-mediated system (Fig. 1). siRNA is a powerful tool for host plants because of its target specificity and rapid efficient response. Plants may have evolved siRNA-mediated regulation as a result of long-term selective pressure, resulting from the struggle with mobile elements.
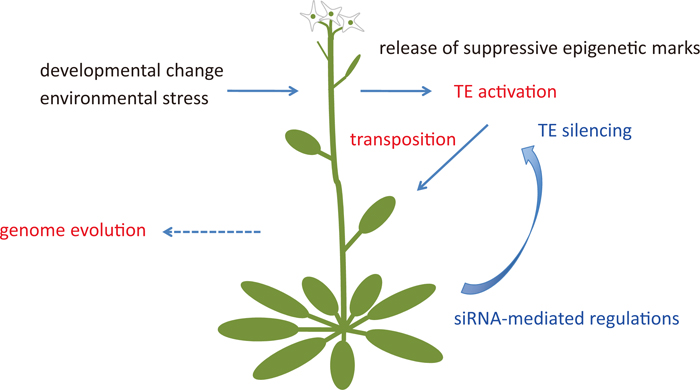
A proposed model for the role of siRNA in regulation of transposable elements (TEs) in plants. Developmental changes or environmental stress affect epigenetic modifications and release suppressive elements such as DNA methylation and histone modification. The removal of epigenetic elements activates TEs and the TEs attempt to transpose and amplify their copy number. The host genome produces siRNAs that regulate TEs by transcriptional suppression and possibly by transpositional inhibition. If a TE escapes regulation and is integrated into a host genome, it may promote genomic evolution of the host plant.
In addition, recent studies have revealed new roles for siRNAs as transgenerational signals that include paramutation and stress memory. Paramutation involves an allelic interaction that leads to heritable changes in gene expression. In maize, alleles in siRNA production impaired mutants lose their paramutagenic future (Alleman et al., 2006). siRNA also related to epigenetic memory. Progeny of a biotic-stressed plant showed higher tolerance to the same stress (Luna et al., 2012; Rasmann et al., 2012). Transgenerational memory, such as a response to biotic and abiotic stress may be transmitted to the next generation via siRNAs.
Although further investigation is necessary to understand the relationship between the observed phenomena and the role of siRNAs, small RNAs play an important role in host genomes in transmitting transgenerational information and facilitating transgenerational regulation of transposons.
I thank Derek Goto for proofreading and his critical comments on the manuscript. This work was supported by JST, PRESTO, a Grant-in-Aid for Scientific Research on Innovative Areas (23119501), a Grant-in-Aid for Young Scientists (B) (23770034), the NIG Cooperative Research Program (2012-B2).