ABSTRACT
Plants subjected to abiotic stress can regulate gene expression post-transcriptionally by means of small RNAs such as microRNAs. Cool-temperature stress causes abnormal tapetum hypertrophy in rice anthers, leading to pollen sterility. As a first step toward understanding the molecular mechanisms of cool tolerance in developing anthers of rice, we report here a comprehensive comparative analysis of microRNAs between cool-sensitive Sasanishiki and cool-tolerant Hitomebore cultivars. High-throughput Illumina sequencing revealed 241 known and 46 novel microRNAs. Interestingly, 15 of these microRNAs accumulated differentially in the two cultivars at the uninucleate microspore stage under cool conditions. Inverse correlations between expression patterns of microRNAs and their target genes were confirmed by quantitative RT-PCR analysis, and cleavage sites of some of the target genes were determined by 5’ RNA ligase-mediated RACE experiments. Thus, our data are useful resources to elucidate microRNA-mediated mechanism(s) of cool tolerance in rice anthers at the booting stage.
INTRODUCTION
The precise development of pollen grains in anthers is achieved by cooperative regulation between gametophyte and sporophyte cells. If such finely balanced cooperation is disorganized under stress, male sterility occurs in plant reproduction (Oda et al., 2010). Male gametophyte development in Oryza sativa L. is susceptible to cool-temperature stress (Sakai, 1937, 1943; Satake and Hayase, 1970). Cool-temperature injury at the booting stage inflicts severe damage to rice pollen. Abnormal tapetum hypertrophy at the tetrad to early uninucleate microspore (UNM) stages is a major cause of pollen sterility, leading to a reduction of the rice grain yield (Sakai, 1949). Cool temperatures in the summer of 1993 in the Tohoku region of Japan caused heavy damage to Japanese rice production: total rice grain yield decreased about 44% in the region, and about 33% in Japan, compared to the average yield for the previous 30 years (MAFF, 2012). Sakai (1949) observed differences in the occurrence of abnormal tapetum between rice cultivars, and showed that cool-tolerant rice cultivars at the booting stage have less abnormal tapetum structure. We have studied two japonica rice cultivars, Hitomebore and Sasanishiki, cultivated in the Tohoku region (Oda et al., 2010). These two cultivars are genetically not distant; however, their response to cool-temperature stress at the booting stage is clearly different. In our previous study, under cool-temperature growth conditions at the booting stage, pollen sterility following incomplete tapetum degradation was observed in Sasanishiki, while anthers from Hitomebore were able to produce normal pollen grains (Oda et al., 2010).
Genetic systems for conferring cool tolerance in rice are not clear, although many quantitative trait loci (QTLs) for cool tolerance at the booting stage have been detected (Takeuchi et al., 2001; Andaya and Mackill, 2003; Ye et al., 2010; Zhou et al., 2010; Kuroi et al., 2011). Saito and his colleagues (2001, 2004, 2010) have explored QTLs that correlated with cold tolerance of spikelet fertility at the booting stage in rice, and they indicated that an F-box protein gene, located on chromosome 4, is one of the genes conferring cold tolerance based on results from map-based cloning. For other QTLs, however, identification of gene(s) has not been completed.
Abiotic stress from environmental changes can severely damage plants even if the stress is short-term. Rapid regulation of transcript level by small RNA is therefore a useful response to the unexpected and unavoidable consequences of abiotic stress. MicroRNAs (miRNAs) are endogenous, approximately 21–24 nucleotides (nt) long, non-coding RNAs belonging to the family of small RNAs, and are thought to play important roles in developmental processes and stress responses by regulating target mRNA expression post-transcriptionally in plants (Bartel, 2004; Kidner and Martienssen, 2005). The primary miRNA (pri-miRNA) that is transcribed by polymerase II from an miRNA gene locus is processed via precursor miRNA into mature miRNA/miRNA* duplex, which has a stem-loop structure, by Dicer-like (DCL) 1 in plants (Kurihara and Watanabe, 2004). The miRNA that is incorporated into RISC (RNA-induced silencing complex) recognizes specific mRNA targets based on fully or partly complementary sequence and guides the RISC to repress target transcripts by translational inhibition and silencing of the target mRNAs (Voinnet, 2009). It has been shown that plants undergoing abiotic stresses regulate the expression of specific genes by miRNAs (Abdel-Ghany and Pilon, 2008; Jeong et al., 2009; Vidal et al., 2010); such regulation was also observed in rice shoots at the prophyll emergence stage (Lv et al., 2010) and in 7-day-old rice seedlings (Yang et al., 2013) under a 4 ℃ treatment.
Although previous papers have suggested a relationship between miRNA profiles and rice anther or pollen development (Wei et al., 2011; Peng et al., 2012; Yan et al., 2015), further investigation is needed to understand the complete picture of the molecular genetic system(s) active in this tissue, and in particular the role of miRNAs in developing rice anthers under cool stress. Here, we focused on miRNA accumulation in immature anthers of rice under cool stress related to cool-temperature injury at the booting stage, and compared the miRNA profiles between cool-sensitive Sasanishiki and cool-tolerant Hitomebore cultivars. Comprehensive analysis of anther miRNA using next-generation sequencing (NGS) showed that some miRNAs are differentially accumulated in UNM anthers under cool stress in the cool-tolerant cultivar. The data obtained in this study are useful resources to understand microRNA-mediated mechanism(s) of cool tolerance in rice anthers at the booting stage.
MATERIALS AND METHODS
Plant materials
Two japonica rice cultivars, Sasanishiki and Hitomebore, were grown in an experimental field at Miyagi Prefectural Furukawa Agricultural Experimental Station, Japan under normal growth conditions. For the cool growth condition, cool-temperature stress was applied using a deep water irrigation system with cool water (19 ℃, 25 cm deep) (Matsunaga, 2005) in the experimental field, according to Oda et al. (2010). Anthers at the UNM and tricellular pollen (TCP) stages were collected and stored at −80 ℃ until analysis.
Illumina sequencing
Total RNA was extracted from rice anthers using the mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA). Each cDNA library was constructed using the TruSeq Small RNA Sample Prep Kit (Illumina, San Diego, CA, USA) with a standard protocol, and single-read multiplex sequencing (read length: 100 bp) was conducted by Illumina Hiseq2000 sequencer (Illumina).
Bioinformatic analysis of sequence data
After filtering low-quality reads and trimming adaptor sequences using Cutadapt (http://code.google.com/p/cutadapt/; Martin, 2011), we collected sequences ranging from 18 to 30 nt. All the collected sequences were mapped to the corresponding cultivar genomes using Bowtie v0.12.9 (Langmead et al., 2009), and sequences that perfectly matched the genome were collected and used for subsequent analysis. Degradation fragments of rRNAs and tRNAs, small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs) were eliminated using blastn (BLAST v2.2.26+) against RAP-DB build 5 (http://rapdblegacy.dna.affrc.go.jp/; Kawahara et al., 2013) and NONCODE v2.0 (http://www.noncode.org/; He et al., 2008), respectively, with an allowance of one mismatch. Sequences that perfectly matched miRNA precursors and overlapped mature rice miRNA sequences from miRBase release 19 (http://www.mirbase.org/; Kozomara and Griffiths-Jones, 2011) were identified as known miRNAs (osa-miRNAs). Sequences that matched rice repeat sequences using blastn against Repbase version 16.12 (http://www.girinst.org/repbase/; Jurka et al., 2005), with an allowance of one mismatch, were identified as repeat-associated small (ra-si)RNAs. Sequences that perfectly matched the TAS3a gene (CI412792) were regarded as trans-acting (ta-si)RNAs.
Among uncategorized sequences, if they were mapped to less than 10 loci in the reference genome, upstream and downstream sequences of the corresponding genomic positions were extracted (the extracted windows were shifted or increased per 10 nt from 50 to 150 bp upstream or downstream), and their secondary structures were predicted using RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi; Hofacker, 2003) with default parameters. Extracted segments were considered as novel miRNAs (nov-miRNAs) if their secondary structure indicated the following characteristics: (1) minimum free energy is less than -18 kcal/mol; (2) the total number of mismatches and gaps between putative miRNA and miRNA* is four or less, but the number of gaps is one or less (Meyers et al., 2008); (3) putative miRNA and miRNA* sequences are in the same library and miRNA counts exceed miRNA* counts; (4) the normalized read count value of a putative miRNA (reads per million, RPM) is over 10 in the library.
The target genes of each miRNA were predicted by the psRNATarget web program (http://plantgrn.noble.org/psRNATarget/; Dai and Zhao, 2011) using rice cDNA sequences from MSU release 7 (http://rice.plantbiology.msu.edu/) with default parameters.
Quantitative reverse transcription (qRT)-PCR
To validate the Illumina sequencing of rice miRNAs, expression levels of miRNAs or target genes were investigated by qRT-PCR. For the target genes, total RNA was extracted from anthers at each developmental stage using the mirVana miRNA Isolation Kit, followed by first-strand cDNA synthesis by Superscript III (Invitrogen, Carlsbad, CA, USA). Small RNA was extracted from the total RNA according to the small RNA isolation protocol (Invitrogen), followed by cDNA synthesis using the Mir-X miRNA first-strand synthesis kit (Clontech, Palo Alto, CA, USA). The reaction mixture (10 μl) consisted of 1×mRQ Buffer, 1.25 μl mRQ Enzyme and 3.75 μl RNA sample, and the mixture was incubated for 1 h at 37 ℃ and then terminated at 85 ℃ for 5 min to inactivate the enzyme. Reaction products were diluted with 90 μl ddH2O. qRT-PCR was performed in an MJ Mini Thermal Cycler and MiniOpticon Real-Time PCR System (Bio-Rad Laboratories, Hercules, CA, USA) using SYBR Advantage qPCR Premix (Clontech). The PCR reaction mixture (25 μl) consisted of 0.2 μM forward and reverse primers, 1×SYBR Advantage qPCR Premix and about 100 ng cDNA, and the PCR cycle parameters were 95 ℃ for 30 sec followed by 45 cycles of 95 ℃ for 5 sec and 60 ℃ for 10 sec. The miRNA forward primers were designed based on the full miRNA sequences, and the mRQ 3’ primer in the Mir-X miRNA first-strand synthesis kit was used as the reverse primer. U6 snRNA was used as the reference gene for miRNA expression, and eukaryotic translation elongation factor 1a (eEF-1a), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), ubiquitin 5 (UBQ5) and U6 snRNA were used as the references for target gene expression. Primers used for qRT-PCR experiments are listed in Supplementary Table S1. The ΔΔCt method was adopted to calculate relative expression value.
5’ RNA ligase-mediated (RLM) RACE
To investigate cleavage sites of predicted target genes, 5’ RLM-RACE was conducted for representative target genes. Approximately 250 ng of mRNA extracted from anthers of Hitomebore at the UNM stage under cool conditions was ligated to a 5’ RACE Adapter, without calf intestine alkaline phosphatase treatment, using a FirstChoice RLM-RACE Kit (Ambion), followed by reverse transcription according to the kit protocol. One microliter of reverse transcription product was used as a template for outer 5’ RLM-RACE PCR using the 5’ RACE Outer primer supplied in the kit and a gene-specific outer primer designed for each putative target gene (Supplementary Table S2). Next, 1 μl Outer PCR product was used as a template for inner 5’ RLM-RACE PCR using the 5’ RACE Inner primer supplied in the kit and a gene-specific inner primer designed for each putative target gene-specific upstream site of the outer gene-specific primer-binding site (Supplementary Table S2). The electrophoresed inner PCR products were purified, and cloned into the pTAKN-2 vector (BioDynamics Laboratory, Tokyo, Japan). More than six independent insert-positive clones were sequenced with a 3500 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) and analyzed by Sequencing Analysis software v.5.4 (Applied Biosystems).
RESULTS AND DISCUSSION
Characteristics of small RNA sequences in rice anthers
Illumina high-throughput sequencing was conducted on eight libraries of small RNAs in rice anthers collected from each of two cultivars (Sasanishiki or Hitomebore) at the UNM or TCP stage under normal or cool growth conditions (Table 1). We selected these eight samples to compare small RNA profiles between the cool-sensitive Sasanishiki and cool-tolerant Hitomebore in immature anthers under cool stress. After removing low-quality data and adaptor sequences, we obtained over 670 million reads in total (Supplementary Table S3). Sequences ranging from 18 to 30 nt were extracted from all the reads and were mapped where possible to the corresponding cultivar genome; the percentage of reads that mapped perfectly to the genome was 96.3 (Supplementary Table S3). Next, we removed rRNAs, tRNAs, snRNAs and snoRNAs from those reads that mapped perfectly to the genome, and finally obtained small RNA sequences comprising ta-siRNAs, ra-siRNAs, known miRNAs (osa-miRNAs), novel miRNAs (nov-miRNAs) and un-annotated RNA (Supplementary Table S3). From an analysis of size distributions, the frequencies of small RNAs were maximum at 21 and 24 nt in all samples (Fig. 1). Under normal growth conditions, sequences of 24 nt were a higher proportion than those of 21 nt, regardless of cultivar and developmental stage (Fig. 1A), while under cool conditions, sequences of 21 nt were a higher proportion than those of 24 nt at the UNM stage in both cultivars (Fig. 1B). Thus, cool-temperature stress appeared to promote the accumulation of 21-nt small RNAs at the UNM stage in rice. This may be a reflection of the stage-specific miRNA expression profile in anthers under cool stress, because the sizes of miRNAs and ta-siRNAs were mainly 21 nt, as determined by DCL1 and DCL4, respectively, in plants (Bartel, 2004; Voinnet, 2009) but ta-siRNAs were rarely detected in any of our libraries compared with the level of miRNA (Supplementary Table S3).
Table 1. Small RNA libraries of rice anther used in this study
| Small RNA library | Cultivar | Developmental
stage | Growth
condition |
|---|
| Sasa-UNM-Normal | Sasanishiki | UNM | Normal |
| Sasa-UNM-Cool | Sasanishiki | UNM | Cool |
| Sasa-TCP-Normal | Sasanishiki | TCP | Normal |
| Sasa-TCP-Cool | Sasanishiki | TCP | Cool |
| Hitome-UNM-Normal | Hitomebore | UNM | Normal |
| Hitome-UNM-Cool | Hitomebore | UNM | Cool |
| Hitome-TCP-Normal | Hitomebore | TCP | Normal |
| Hitome-TCP-Cool | Hitomebore | TCP | Cool |

To annotate osa-miRNAs, the reads of each library were mapped to the precursor and mature sequences from the rice miRNA database available in miRBase (release 19). As a result, a total of 3,765,844 reads were mapped to the osa-miRNAs (Supplementary Table S3), and 239 out of 289 miRNA families available in the miRBase were detected in rice anthers in this study. Furthermore, 48 miRNA families were regarded as nov-miRNAs (Table 2), according to the criteria described in MATERIALS AND METHODS. After this analysis, we checked a more recent version of miRBase (release 21), and found that two of the 48 nov-miRNAs are now annotated as osa-miRNAs. Thus, we finally identified 241 osa-miRNAs and 46 nov-miRNAs in this study.
Table 2. Novel miRNAs detected in developing rice anthers in this study
| miRNA | miRNA sequence | nt |
|---|
| nov-miR001 | AAGGAUGACUUGCCGAUCGAUCGA | 24 |
| nov-miR002 | ACACCGAUAUGCGUCAUCAUCAAG | 24 |
| nov-miR003 | ACCUAGGACAAGGACAUGGCAUCC | 24 |
| nov-miR004 | AGAAAAAAACUUUAGCGCCUUGAC | 24 |
| nov-miR005 | AGAAGAGAGAGAGUACAGCCU | 21 |
| nov-miR006 | AGAUUCGUAGUACUAGAAUGUAUU | 24 |
| nov-miR007 | AGGAUUAGAGGGAACUGAACC | 21 |
| nov-miR008 | AGGUCAAUCGUUGUUGCAAGUUCU | 24 |
| nov-miR009 | AUCAGUUCGUAUUUUGUGGAC | 21 |
| nov-miR010 | AUCGGAGGCCAUGGUGCAGCC | 21 |
| nov-miR011 | CAUGUCUGUUUCUACUAAG | 19 |
| nov-miR012 | CAUGUGCCCUUCUUCUCCACC | 21 |
| nov-miR013 | CCAGUAGAUAGUAUAUAUAGU | 21 |
| nov-miR014 | CCAUCCUAGGACUACGAAUCUGGA | 24 |
| nov-miR015 | CGACGUAUAUCGGUGUUCGCU | 21 |
| nov-miR016 | CGCUAUAUCGAGAGGAAGGUAGUA | 24 |
| nov-miR017 | CGGAUGGUUCUUCUGCCU | 18 |
| nov-miR018 | CGGCAUAAUUAGAUCACUUGAU | 22 |
| nov-miR019 | CGUGGAAGAUUUCUCCGUUCGCCU | 24 |
| nov-miR020 | CUAUAUAUACUAUCUACUGGU | 21 |
| nov-miR021 | CUUGAGCUGAGCUUCCCGUGGA | 22 |
| nov-miR022 | GAUGUACUUUUUGUUGGACC | 20 |
| nov-miR023 | GAUGUCGCAGAUGGCCGUGCU | 21 |
| nov-miR024 | GCUCGAGCGUCCCUGCAC | 18 |
| nov-miR025 | GCUGUUGUCGUCAAGGCGCUAAAG | 24 |
| nov-miR026 | GCUGUUGUCGUCGAGGCGCUAAAG | 24 |
| nov-miR027 | GCUGUUGUCGUCGAUGCGCUAAAG | 24 |
| nov-miR028 | GGCGUGCCGAUGAUCGCCGGCA | 22 |
| nov-miR029 | GGUUCCUGUCCCAAGACUGA | 20 |
| nov-miR030 | GUAUGAGCCCCGUGGACUAGC | 21 |
| nov-miR031 | GUAUUAGAAUAUGUCGCAUCCG | 22 |
| nov-miR032 a | GUGCUUUCUCCCGUUGUCACU | 21 |
| nov-miR033 | GUUAGAUUGGCUUGGAGUGGU | 21 |
| nov-miR034 | GUUUCCGCGCCCAACUUUGAUCGU | 24 |
| nov-miR035 | GUUUCCGCGUCCAACUUUGAUCGU | 24 |
| nov-miR036 | GUUUGUGUUCUUAGAAACUCGGGA | 24 |
| nov-miR037 | GUUUUCGCACCCAACUUUGAUCGU | 24 |
| nov-miR038 | GUUUUCGUGUCCAACUUUGACUGU | 24 |
| nov-miR039 | UACUGUAACUUUUAGCUUGGC | 21 |
| nov-miR040 | UCACUUGAUGACGACGUAUAUC | 22 |
| nov-miR041 | UCAGUACGACCAAUAGAACCAU | 22 |
| nov-miR042 | UCCGGAGUUUCUAAGAACACAAAC | 24 |
| nov-miR043 | UCCUAGGACAAUGUAUGUAGACAU | 24 |
| nov-miR044 | UCGGAGGUGACUCACUUGUGA | 21 |
| nov-miR045 | UGAUACCAAUUGUCGGCACUC | 21 |
| nov-miR046 | UGUAGAGAUUAAUCAUUAGACGAG | 24 |
| nov-miR047 | UUUGGCUUGCAACGUUUGACC | 21 |
| nov-miR048 b | UUUGUUGCAAUUUGGACUACC | 21 |
a assigned as osa-miR535-3p in miRBase release 21.
b assigned as osa-miR5827 in miRBase release 21.
The raw count of each osa-miRNA and nov-miRNA was normalized to RPM. We selected 110 miRNAs whose expression levels were over 10 RPM in at least one of the eight samples for further comparative analysis between Sasanishiki and Hitomebore. The 110 miRNAs were detected in rice anthers under both normal and cool conditions; neither normal-condition-specific nor cool-condition-specific miRNAs were included in this set of miRNAs. Under cool conditions, 89.1% of the miRNAs were common among the two stages (UNM and TCP) and the two cultivars (Fig. 2A). At the UNM anther stage under cool conditions, most of the miRNAs were expressed at a higher level in Hitomebore than Sasanishiki (Fig. 2B), although this difference was not observed in the TCP stage (Fig. 2C). These data suggest that the differential accumulation of miRNAs between Sasanishiki and Hitomebore effects cool tolerance of rice anthers at the UNM stage, when the tapetum cells are being degraded (Oda et al., 2010). We therefore focused on the expression level of miRNAs at the UNM stage to explore miRNA(s) related to cool tolerance in rice in this study.

We selected miRNAs differentially expressed between Sasanishiki and Hitomebore at the UNM stage under the cool condition that satisfied both of the following conditions: 1) a value of more than 1 RPM was detected in both the cultivars, and 2) the RPM value differed by more than two-fold between the cultivars. We thus identified 13 miRNAs that were differentially expressed: seven osa-miRNAs (osa-miR397b, osa-miR398b, osa-miR408-3p, osa-miR528-5p, osa-miR171b/c/d/e/f, osa-miR1432-5p and osa-miR5072) and six nov-miRNAs (nov-miR017, nov-miR018, nov-miR023, nov-miR024, nov-miR030 and nov-miR031) (Table 3). In addition, we identified two cultivar-specific miRNAs (osa-miR531a/c and nov-miR039) that accumulated to more than 10 RPM in one cultivar and less than 1 RPM in the other cultivar; osa-miR531 and nov-miR039 were specifically expressed in Sasanishiki and Hitomebore, respectively (Table 3). It is noteworthy that seven osa-miRNAs, out of the 13 differentially expressed and two cultivar-specific miRNAs, have previously been reported to be expressed in developing rice pollen (Wei et al., 2011). Thus, we considered that these 15 miRNAs were candidates for rice miRNAs related to cool tolerance in immature anthers.
Table 3. Fifteen miRNAs expressed differentially between Sasanishiki and Hitomebore under cool growth conditions
| miRNA | Sequence | RPM | RPM log2
(cool/normal ratio)
at the UNM stage |
|---|
Sasa-
UNM-
Normal | Sasa-
UNM-
Cool | Sasa-
TCP-
Normal | Sasa-
TCP-
Cool | Hitome-
UNM-
Normal | Hitome-
UNM-
Cool | Hitome-
TCP-
Normal | Hitome-
TCP-
Cool | Sasanishiki | Hitomebore |
|---|
differentially
expressed | osa-miR397b | UUGAGUGCAGCGUUGAUGAACC | 53.1 | 62.9 | 258.9 | 215.6 | 25.7 | 273.6 | 308.0 | 268.6 | 0.24 | 3.42 |
| osa-miR398b | UGUGUUCUCAGGUCGCCCCUG | 8.7 | 19.6 | 10.2 | 13.3 | 4.7 | 80.0 | 1.5 | 11.9 | 1.18 | 4.08 |
| osa-miR408-3p | CUGCACUGCCUCUUCCCUGGC | 27.8 | 50.4 | 74.1 | 73.6 | 16.8 | 133.7 | 18.4 | 63.7 | 0.86 | 2.99 |
| osa-miR528-5p | UGGAAGGGGCAUGCAGAGGAG | 12.5 | 11.1 | 9.7 | 4.1 | 9.2 | 38.3 | 3.9 | 5.9 | –0.17 | 2.06 |
| osa-miR171b/c | UUGAGCCGUGCCAAUAUCACG | 5.1 | 6.9 | 1.2 | 5.9 | 10.5 | 16.2 | 2.3 | 4.1 | 0.42 | 0.63 |
| osa-miR1432-5p | AUCAGGAGAGAUGACACCGACA | 0.3 | 4.2 | 0.1 | 3.0 | 2.9 | 15.3 | 0.1 | 6.1 | 3.82 | 2.39 |
| osa-miR5072 | UCCCCAGCGGAGUCGCCA | 7.0 | 14.6 | 5.2 | 7.3 | 8.7 | 5.7 | 5.1 | 6.5 | 1.06 | –0.61 |
| nov-miR017 | CGGAUGGUUCUUCUGCCU | 9.4 | 4.8 | 0.3 | 0.6 | 19.3 | 12.0 | 0.1 | 0.7 | –0.96 | –0.68 |
| nov-miR018 | CGGCAUAAUUAGAUCACUUGAU | 4.5 | 1.7 | 12.7 | 28.1 | 1.8 | 4.7 | 73.9 | 38.1 | –1.39 | 1.41 |
| nov-miR023 | GAUGUCGCAGAUGGCCGUGCU | 2.4 | 4.0 | 0.1 | 0.3 | 10.8 | 10.4 | 0.2 | 0.5 | 0.73 | –0.05 |
| nov-miR024 | GCUCGAGCGUCCCUGCAC | 12.0 | 2.3 | 1.9 | 1.0 | 12.1 | 5.9 | 1.0 | 1.0 | –2.37 | –1.05 |
| nov-miR030 | GUAUGAGCCCCGUGGACUAGC | 15.6 | 2.4 | 4.5 | 5.0 | 49.6 | 5.1 | 2.5 | 3.8 | –2.73 | –3.28 |
| nov-miR031 | GUAUUAGAAUAUGUCGCAUCCG | 8.1 | 5.6 | 0.2 | 0.5 | 5.8 | 16.2 | 0.1 | 0.8 | –0.53 | 1.47 |
cultivar-specifically
expressed | osa-miR531a/c | CUCGCCGGGGCUGCGUGCCGCC | 5.6 | 16.7 | 18.6 | 16.7 | ND a | ND | ND | ND | 1.58 | – |
| nov-miR039 | UACUGUAACUUUUAGCUUGGC | 0.6 | 0.5 | 0.03 | 0.3 | 33.6 | 48.3 | 20.8 | 22.1 | –0.19 | 0.52 |
For the 15 candidate miRNAs, we calculated the RPM ratios between different growth conditions (cool/normal) to understand the up- or down-regulation of their expression under cool stress (Fig. 3). In Sasanishiki, four miRNAs (osa-miR398b, osa-miR1432-5p, osa-miR5072 and osa-miR531a/c) were up-regulated over two-fold by cool-temperature stress (Fig. 3A). In Hitomebore, seven miRNAs (osa-miR397b, osa-miR398b, osa-miR408-3p, osa-miR528-5p, osa-miR1432-5p, nov-miR018 and nov-miR031) were up-regulated over two-fold by cool-temperature stress (Fig. 3B). Most of the up-regulated miRNAs showed a greater change of RPM in Hitomebore than in Sasanishiki in response to cool stress (Fig. 3), indicating a greater influence of cool-temperature stress in Hitomebore than in Sasanishiki. Such differences in miRNA profiles between stress-sensitive and stress-tolerant genotypes have been observed in other plants subjected to abiotic or biotic stress (drought stress in cowpea, Barrera-Figueroa et al., 2011; salt stress in maize, Ding et al., 2009; drought or disease stress in soybean, Kulcheski et al., 2011). These examples suggest that plant miRNAs play an important role in stress tolerance.

To validate the miRNA expression profiles obtained using Illumina sequencing, qRT-PCR was conducted against five representative miRNAs (osa-miR397b, osa-miR398b, osa-miR408-3p, osa-miR528-5p and nov-miR018) (Fig. 4), which were selected as miRNAs showing a high cool/normal temperature RPM ratio in Hitomebore (Fig. 3B). The expression levels and patterns of miRNAs in the qRT-PCR results correlated almost exactly with those from Illumina sequencing, although a clear difference between cultivars was not detected in nov-miR018 expression (Fig. 4, I and J).
Prediction of target genes related to cool tolerance in rice at the booting stage
Analysis of miRNAs enables computational prediction of miRNA targets because plant miRNAs have almost perfectly complementary sequences to their target mRNAs. Therefore, we predicted the target genes of the 15 candidate miRNAs using the psRNATarget web program with default parameters. In total, 64 target genes were predicted for 12 of the miRNAs, and these were categorized into 34 gene products (Table 4). No target genes were predicted for the other three miRNAs. Interestingly, the most frequent annotations were related to redox genes, with 20 target genes being redox-related: two genes (LOC_Os07g46990 and LOC_Os08g44770) targeted by osa-miR398b and osa-miR528-5p encode copper/zinc superoxide dismutase (SOD); 14 genes (e.g., LOC_Os01g44330) targeted by osa-miR397b and osa-miR528-5p encode laccase precursor; LOC_Os06g37150 targeted by osa-miR528-5p encodes L-ascorbate oxidase precursor; two genes (LOC_Os01g03620 and LOC_Os01g03640) targeted by osa-miR528-5p contain a multicopper oxidase domain; and LOC_Os08g39960 targeted by osa-miR531 encodes 3-oxoacyl-reductase (Table 4). Of the 15 miRNAs, miR171c, miR398b and miR408 are highly conserved in Arabidopsis (Sunker and Zhu, 2004), and miRNA-target regulatory modules in Arabidopsis were also predicted in this study, such as miR171 and its target genes (scarecrow-like family genes) (Llave et al., 2002), miR398 and its target genes (copper/zinc SOD genes, CSD1 and CSD2) (Sunkar et al., 2006), and miR408 and its target gene (plastocyanin) (Abdel-Ghany and Pilon, 2008). These data suggest that our predictions of target genes show good reliability, and these miRNA-target gene properties may be conserved between plant species.
Table 4. Putative genes targeted by 12 miRNAs expressed differentially in anthers of two rice cultivars at the UNM stage under cool growth conditions
| miRNA family | Target MSU accession | Target gene description |
|---|
| osa-miR397b | LOC_Os01g44330 | laccase precursor protein, putative |
| LOC_Os01g62480 | laccase precursor protein, putative |
| LOC_Os01g62490 | laccase precursor protein, putative |
| LOC_Os01g63180 | laccase-6 precursor, putative |
| LOC_Os01g63190 | laccase precursor protein, putative |
| LOC_Os01g63200 | laccase precursor protein, putative |
| LOC_Os02g34600 | CAMK_CAMK_like.13 - CAMK includes calcium/calmodulin depedent protein kinases |
| LOC_Os03g16610 | laccase precursor protein, putative |
| LOC_Os05g35770 | serine/threonine-protein kinase SAPK4, putative |
| LOC_Os05g38410 | laccase precursor protein, putative |
| LOC_Os05g38420 | laccase precursor protein, putative |
| LOC_Os11g01730 | laccase-23 precursor, putative |
| LOC_Os11g48060 | laccase-22 precursor, putative |
| LOC_Os12g01730 | laccase-23 precursor, putative |
| LOC_Os12g15530 | retrotransposon protein, putative, unclassified |
| LOC_Os12g15680 | laccase precursor protein, putative |
| osa-miR398b | LOC_Os07g46990 | copper/zinc superoxide dismutase, putative |
| LOC_Os11g09780 | expressed protein |
| osa-miR408-3p | LOC_Os02g43660 | plastocyanin-like domain containing protein, putative |
| LOC_Os03g05650 | expressed protein |
| LOC_Os10g41040 | ternary complex factor MIP1, putative |
| LOC_Os11g01470 | retrotransposon protein, putative, unclassified |
| ChrSy.fgenesh.mRNA.77 | expressed protein |
| osa-miR528-5p | LOC_Os01g03620 | multicopper oxidase domain containing protein |
| LOC_Os01g03640 | multicopper oxidase domain containing protein |
| LOC_Os01g40150 | eukaryotic translation initiation factor 5B, putative |
| LOC_Os01g44330 | laccase precursor protein, putative |
| LOC_Os06g06050 | OsFBL27 - F-box domain and LRR containing protein |
| LOC_Os06g37150 | L-ascorbate oxidase precursor, putative |
| LOC_Os07g38290 | plastocyanin-like domain containing protein, putative |
| LOC_Os08g04310 | plastocyanin-like domain containing protein, putative |
| LOC_Os08g44770 | copper/zinc superoxide dismutase, putative |
| LOC_Os09g33800 | arabinogalactan protein, putative |
| LOC_Os12g13295 | expressed protein |
| osa-miR171b/c | LOC_Os02g44360 | scarecrow transcription factor family protein, putative |
| LOC_Os02g44370 | scarecrow, putative |
| LOC_Os04g42000 | 6,7-dimethyl-8-ribityllumazine synthase, chloroplast precursor, putative |
| LOC_Os04g46860 | scarecrow, putative |
| LOC_Os06g01620 | scarecrow, putative |
| LOC_Os10g21360 | N-dimethylguanosine tRNA methyltransferase, putative |
| osa-miR1432-5p | LOC_Os03g59770 | EF hand family protein, putative |
| LOC_Os03g59790 | EF hand family protein, putative |
| LOC_Os04g35590 | thioesterase family protein, putative |
| LOC_Os05g07210 | metal cation transporter, putative |
| osa-miR531 | LOC_Os02g20934 | expressed protein |
| LOC_Os08g39960 | 3-oxoacyl-reductase, chloroplast precursor, putative |
| LOC_Os09g27734 | expressed protein |
| LOC_Os12g19530 | ATP-binding region, ATPase-like domain-containing protein, putative |
| nov-miR018 | LOC_Os02g20970 | C2 domain containing protein, putative |
| LOC_Os06g24711 | expressed protein |
| nov-miR023 | LOC_Os01g66690 | ZIP4/SPO22, putative |
| LOC_Os02g13410 | leucine-rich repeat family protein, putative |
| LOC_Os04g39660 | OsFBDUF22 - F-box and DUF domain containing protein |
| LOC_Os05g44570 | histidine-containing phosphotransfer protein, putative |
| LOC_Os08g17990 | retrotransposon protein, putative, unclassified |
| nov-miR030 | LOC_Os05g30680 | leucine zipper protein-like, putative |
| LOC_Os06g35900 | BES1/BZR1 homolog protein, putative |
| LOC_Os12g02280 | pentatricopeptide, putative |
| nov-miR031 | LOC_Os01g41240 | hydrolase, alpha/beta fold family domain containing protein |
| LOC_Os03g51230 | SNF2 family N-terminal domain containing protein |
| LOC_Os04g32540 | OsSCP24 - Putative Serine Carboxypeptidase homologue |
| LOC_Os04g40970 | DEAD-box ATP-dependent RNA helicase, putative |
| LOC_Os04g59624 | SNF2 family N-terminal domain containing protein |
| nov-miR039 | LOC_Os01g71106 | NBS-LRR disease resistance protein, putative |
We investigated expression levels and patterns of four representative predicted target genes (LOC_Os01g03640, LOC_Os02g20970, LOC_Os07g46990 and LOC_Os08g44770) by qRT-PCR, and obtained clear inverse correlations between expression levels of the target genes and those of their corresponding miRNAs (Fig. 5). Furthermore, we determined the cleavage sites of six predicted target genes using 5’ RLM-RACE analysis (Supplementary Table S2; Fig. 6). Of the six target genes investigated, major cleavage sites of four target mRNAs (LOC_Os07g46990, LOC_Os07g38290, LOC_Os06g06050 and LOC_Os06g37150) were mapped between nucleotides 10 and 11 (counting from the 5’ end of each miRNA) (Fig. 6). However, we obtained unexpected results for the remaining two target genes: 14 of 20 clones from LOC_Os02g44360 were cleaved between nucleotides 9 and 10 (Fig. 6E), and all 22 clones from LOC_Os01g71106 were cleaved between nucleotides 6 and 7 (Fig. 6F). Although we have no explanation for these slippages from canonical cleavage sites, Wei et al. (2011) observed a similar phenomenon in developing rice pollen, suggesting that this may be due to subtle differences in regulatory mechanisms between tissues. Thus, using qRT-PCR and 5’ RLM-RACE techniques, we validated the computational prediction of target genes of miRNAs related to cool tolerance in rice at the booting stage.
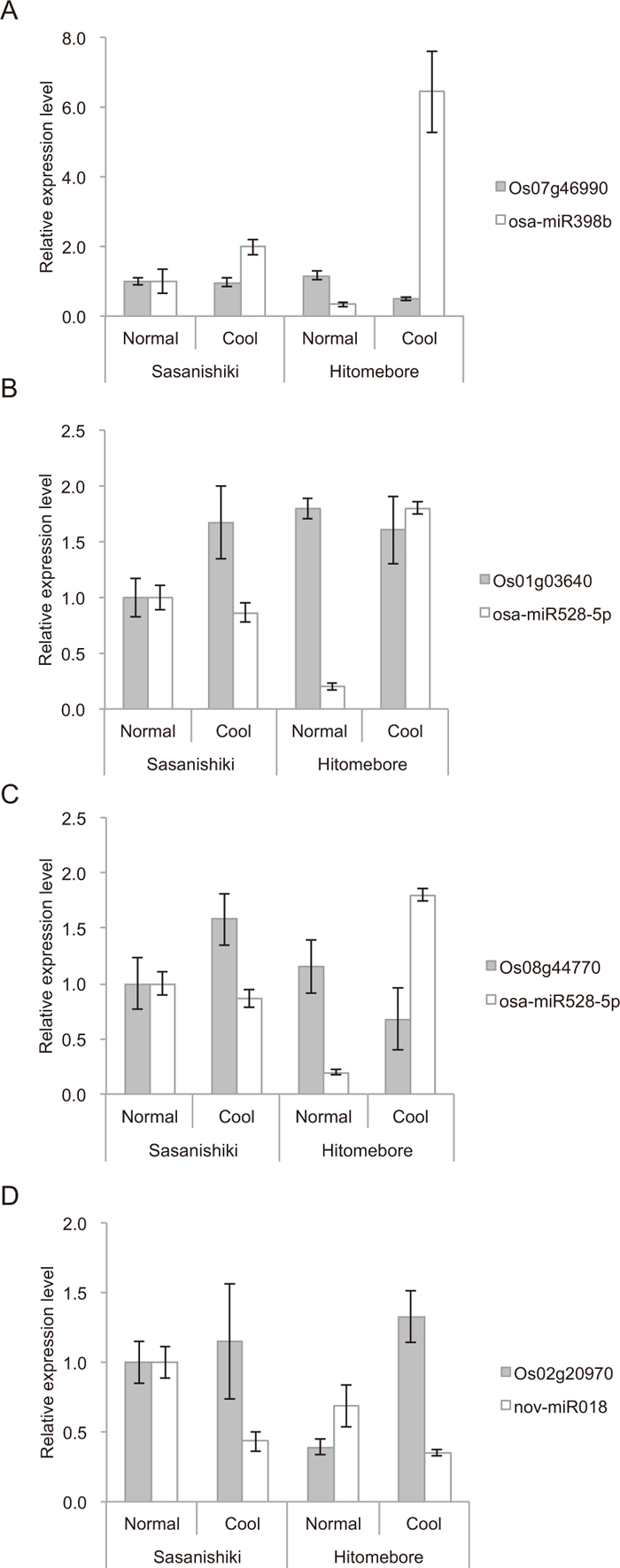
Tapetum cells in rice anthers are degraded by programmed cell death (PCD) during tetrad to vacuolated pollen stages (Li et al., 2006), and retarded degradation of the tapetum can cause pollen sterility (Li et al., 2006; Li et al., 2015). Pollen sterility in rice plants due to cool-temperature stress is mainly due to tapetum hypertrophy (Imin et al., 2006; Oda et al., 2010). Tapetum degradation, therefore, is essential for normal male gametophyte development in rice. Reactive oxygen species (ROS) accumulate in the tapetum during young microspore formation (Hu et al., 2011), and ROS accumulation is involved in tapetum degradation by PCD (Li et al., 2006). Thus, it is possible that SOD, one of the ROS scavengers (Mittler et al., 2004), plays a crucial role in the induction of male sterility under cool stress by disturbing PCD of tapetum cells in rice anthers. In this study, differential expression of osa-miR398b was observed between cool-sensitive and cool-tolerant rice cultivars, and the sequence of osa-miR398b indicates that it could target two SOD genes (LOC_Os07g46990 and LOC_Os08g44770) (Table 4). In Arabidopsis, SOD is regulated at the post-transcriptional level by miR398 (Sunkar et al., 2006; Beauclair et al., 2010); likewise, we clarified that one SOD (LOC_Os07g46990) was down-regulated by osa-miR398b accumulation at the UNM stage under cool-temperature conditions in cool-tolerant Hitomebore (Fig. 5A). We also showed that this SOD (LOC_Os07g46990) transcript can be cleaved as a target of osa-miR398b (Fig. 6A). Similarly, accumulation of osa-miR528-5p may down-regulate the other SOD gene, LOC_Os08g44770 (Fig. 5C). These results suggest that inhibition of SOD by osa-miR398b and osa-miR528-5p contributes to the maintenance of normal tapetum degradation and subsequent pollen maturation in Hitomebore under cool stress. The involvement of miRNA-mediated down-regulation of SOD genes in cool tolerance in rice at the booting stage will be elucidated in the future by functional analyses of the candidate miRNAs and their targets.
Conclusion
Using NGS technology, we conducted a comprehensive comparative analysis of miRNAs in rice anthers under cool conditions from the cool-sensitive Sasanishiki and cool-tolerant Hitomebore cultivars. The miRNA profiles reported here are useful resources to deepen our understanding of the regulation of gene expression in plants under abiotic stress. The present study showed that a limited number of miRNAs are differentially expressed between the cultivars and are up-regulated under cool stress at the UNM stage, indicating that small RNA-mediated silencing is involved in the cool-stress response in rice anthers. These miRNAs and their targets are candidates for explaining the basis of the cool-tolerance trait in Hitomebore anthers.
ACKNOWLEDGMENTS
The authors thank Masumi Miyano, Reina Shida, Ayumi Sadaike, Mayumi Maruoka, Tatsuya Kohata, Takuya Ogura and Mizuho Kanamori (Tohoku University) for technical assistance. The authors are also grateful to Dr. Ryohei Terauchi (Iwate Biotechnology Research Center) for providing the genome sequence of two rice cultivars (Hitomebore and Sasanishiki). This work was supported in part by Grants-in-Aid for Scientific Research on Innovative Areas (Nos. 23113006, 23113001 to G.S., K.S., and M.W.), Scientific Research (A) (No. 25252001 to M.W.), Scientific Research (B) (No. 25292005 to K.S.) and Scientific Research (C) (No. 25450515 to G.S.) from the Japan Society for the Promotion of Science (JSPS). M.O. is recipient of a Research Fellowship for Young Scientists from JSPS.
REFERENCES
- Abdel-Ghany, S. E., and Pilon, M. (2008) MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 283, 15932–15945.
- Andaya, V. C., and Mackill, D. J. (2003) QTLs conferring cold tolerance at the booting stage of rice using recombinant inbred lines from a japonica x indica cross. Theor. Appl. Genet. 106, 1084–1090.
- Barrera-Figueroa, B. E., Gao, L., Diop, N. N., Wu, Z., Ehlers, J. D., Roberts, P. A., Close, T. J., Zhu, J-K., and Liu, R. (2011) Identification and comparative analysis of drought-associated microRNAs in two cowpea genotypes. BMC Plant Biol. 11, 127.
- Bartel, D. P. (2004) MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281–297.
- Beauclair, L., Yu, A., and Bouché, N. (2010) microRNA-directed cleavage and translational repression of the copper chaperone for superoxide dismutase mRNA in Arabidopsis. Plant J. 62, 454–462.
- Dai, X., and Zhao, P. X. (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res., 39 (suppl 2), W155–W159.
- Ding, D., Zhang, L., Wang, H., Liu, Z., Zhang, Z., and Zheng, Y. (2009) Differential expression of miRNAs in response to salt stress in maize roots. Ann. Bot. 103, 29–38.
- He, S., Liu, C., Skogerbo, G., Zhao, H., Wang, J., Liu, T., Bai, B., Zhao, Y., and Chen, R. (2008) NONCODE v2.0: decoding the non-coding. Nucleic Acids Res. 36 (suppl 1), D170–D172.
- Hofacker, I. L. (2003) Vienna RNA secondary structure server. Nucleic Acids Res. 31, 3429–3431.
- Hu, L., Liang, W., Yin, C., Cui, X., Zong, J., Wang, X., Hu, J., and Zhang, D. (2011) Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell 23, 515–533.
- Imin, N., Kerim, T., Weinman, J. J., and Rolfe, B. G. (2006) Low temperature treatment at the young microspore stage induces protein changes in rice anthers. Mol. Cell. Proteomics 5, 274–292.
- Jeong, D-H., German, M. A., Rymarquis, L. A., Thatcher, S. R., and Green, P. J. (2009) Abiotic stress-associated miRNAs: Detection and functional analysis. Methods Mol. Biol. 592, 203–230.
- Jurka, J., Kapitonov, V. V., Pavlicek, A., Klonowski, P., Kohany, O., and Walichiewicz, J. (2005) Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110, 462–467.
- Kawahara, Y., de la Bastide, M., Hamilton, J. P., Kanamori, H., McCombie, W. R., Ouyang, S., Schwartz, D. C., Tanaka, T., Wu, J., Zhou, S., et al. (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6, 4.
- Kidner, C. A., and Martienssen, R. A. (2005) The development role of microRNA in plants. Curr. Opin. Plant Biol. 8, 38–44.
- Kozomara, A., and Griffiths-Jones, S. (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39 (suppl 1), D152–D157.
- Kulcheski, F. R., de Oliveira, L. F. V., Molina, L. G., Almerão, M. P., Rodrigues, F. A., Marcolino, J., Barbosa, J. F., Stolf-Moreira, R., Nepomuceno, A. L., Marcelino-Guimarães, F. C., et al. (2011) Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genomics 12, 307.
- Kurihara, Y., and Watanabe, Y. (2004) Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Arad. Sci. USA 101, 12753–12758.
- Kuroi, M., Saito, K., Matsuba, S., Yokogami, N., Ando, T., Sato, Y., Ando, I., and Shimizu, H. (2011) Detection of quantitative trait loci for cold tolerance at the booting stage in a rice breeding line, Hokkai-PL9. Breed. Res. 13, 11–18.
- Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25.
- Li, N., Zhang, D-S., Liu, H-S., Yin, C-S., Li, X-X., Liang, W-q., Yuan, Z., Xu, B., Chu, H-W., Wang, J., et al. (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18, 2999–3014.
- Li, L., Li, Y., Song, S., Deng, H., Li, N., Fu, X., Chen, G., and Yuan, L. (2015) An anther development F-box (ADF) protein regulated by tapetum degeneration retardation (TDR) controls rice anther development. Planta 241, 157–166.
- Llave, C., Xie, Z., Kasschau, K. D., and Carrington, J. C. (2002) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056.
- Lv, D-K., Bai, X., Li, Y., Ding, X-D., Ge, Y., Cai, H., Ji, W., Wu, N., and Zhu, Y-M. (2010) Profiling of cold-stress-responsive miRNAs in rice by microarrays. Gene 459, 39–47.
- MAFF (2012) Statistics of Agriculture, Forestry and Fisheries (http://www.e-stat.go.jp/SG1/estat/List.do?bid=000001024932 &cycode=0) (accessed July 2015), Ministry of Agriculture, Forestry and Fisheries (MAFF), Crop Statistics Section, Tokyo, Japan.
- Martin, M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12.
- Matsunaga, K. (2005) Establishment of an evaluation method for cold tolerance at the booting stage of rice using deep water irrigation system and development of highly cool-tolerant rice varieties by combining cold tolerance genes. Bull. Miyagi Furukawa Agric. Exp. Sta. 4, 1–78.
- Meyers, B. C., Axtell, M. J., Bartel, B., Bartel, D. P., Baulcombe, D., Bowman, J. L., Cao, X., Carrington, J. C., Chen, X., Green, P. J., et al. (2008) Criteria for annotation of plant microRNAs. Plant Cell 20, 3186–3190.
- Mittler, R., Vanderauwera, S., Gollery, M., and van Breusegem, F. (2004) Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498.
- Oda, S., Kaneko, F., Yano, K., Fujioka, T., Masuko, H., Park, J-I., Kikuchi, S., Hamada, K., Endo, M., Nagano, K., et al. (2010) Morphological and gene expression analysis under cool temperature conditions in rice anther development. Genes Genet. Syst. 85, 107–120.
- Peng, H., Chun, J., Ai, T-b., Tong, Y-a., Zhang, R., Zhao, M-m., Chen, F., and Wang, S-h. (2012) MicroRNA profiles and their control of male gametophyte development in rice. Plant Mol. Biol. 80, 85–102.
- Saito, K., Miura, K., Nagano, K., Hayano-Saito, Y., Araki, H., and Kato, A. (2001) Identification of two closely linked quantitative trait loci for cold-tolerance on chromosome 4 of rice and their association with anther length. Theor. Appl. Genet. 103, 862–868.
- Saito, K., Hayano-Saito, Y., Maruyama-Funatsuki, W., Sato, Y., and Kato, A. (2004) Physical mapping and putative candidate gene identification of a quantitative trait locus Ctb1 for cold-tolerance at the booting stage of rice. Theor. Appl. Genet. 109, 515–522.
- Saito, K., Hayano-Saito, Y., Kuroki, M., and Sato, Y. (2010) Map-based cloning of the rice cold tolerance gene Ctb1. Plant Sci. 179, 97–102.
- Sakai, K. (1937) Interfering effects of low temperatures upon the microsporogenetic cell division in rice plant. Proc. Crop Sci. Soc. Japan 9, 207–212.
- Sakai, K. (1943) Cyto-histological investigations on the sterility mechanism in rice plants in Hokkaido caused by cold injury in 1941. Rep. Agric. Res. St. Hokkaido 40, 1–17.
- Sakai, K. (1949) Cytological and genetical studies on the sterility of rice plants as influenced by low temperature with special reference to the hypertrophy of tapetum. Rep. Agric. Res. St. Hokkaido 43, 1–46.
- Satake, T., and Hayase, H. (1970) Male sterility caused by cooling treatment at the young microspore stage in rice plants. V. Estimations of pollen developmental stage and the most sensitive stage to coolness. Proc. Crop Sci. Soc. Japan 39, 468–473.
- Sunkar, R., Kappor, A., and Zhu, J-K. (2006) Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18, 2051–2065.
- Sunker, R., and Zhu, J-K. (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16, 2001–2019.
- Takeuchi, Y., Hayasaka, H., Chiba, B., Tanaka, I., Shimano, T., Yamagishi, M., Nagano, K., Sasaki, T., and Yano, M. (2001) Mapping quantitative trait loci controlling cool-temperature tolerance at booting stage in temperate japonica rice. Breed. Sci. 51, 191–197.
- Vidal, E. A., Araus, V., Lu, C., Parry, G., Green, P. J., Coruzzi, G. M., and Gutiérrez, R. A. (2010) Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107, 4477–4482.
- Voinnet, O. (2009) Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687.
- Wei, L. Q., Yan, L. F., and Wang, T. (2011) Deep sequencing on genome-wide scale reveals the unique composition and expression patterns of microRNAs in developing pollen of Oryza sativa. Genome Biol. 12, R53.
- Yan, J., Zhang, H., Zheng, Y., and Ding, Y. (2015) Comparative expression profiling of miRNAs between the cytoplasmic male sterile line MeixiangA and its maintainer line MeixiangB during rice anther development. Planta 241, 109–123.
- Yang, C., Li, D., Mao, D., Liu, X., Ji, C., Li, X., Zhao, X., Cheng, Z., Chen, C., and Zhu, L. (2013) Overexpression of mircoRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ. 36, 2207–2218.
- Ye, C., Fukai, S., Godwin, I. D., Koh, H., Reinke, R., Zhou, Y., Lambrides, C., Jiang, W., Snell, P., and Redoña, E. (2010) A QTL controlling low temperature induced spikelet sterility at booting stage in rice. Euphytica 176, 291–301.
- Zhou, L., Zeng, Y., Zheng, W., Tang, B., Yang, S., Zhang, H., Li, J., and Li, Z. (2010) Fine mapping a QTL qCTB7 for cold tolerance at the booting stage on rice chromosome 7 using a near-isogenic line. Theor. Appl. Genet. 121, 895–905.







