2017 Volume 92 Issue 3 Pages 127-133
2017 Volume 92 Issue 3 Pages 127-133
A chromosome is composed of structurally and functionally distinct domains. Telomeres, which are located at the ends of linear chromosomes, play crucial roles in genome stability. Although substantial knowledge of telomeres has been accumulated, the regulation and function of subtelomeres, which are the domains adjacent to telomeres, remain largely unknown. In this review, I describe recent discoveries about the multiple roles of a shugoshin family protein, Sgo2, which is localized at centromeres in mitosis and contributes to precise chromosome segregation, in defining chromatin structure and functions of the subtelomeres in fission yeast. Sgo2 becomes enriched at the subtelomeres, particularly during G2 phase, and is essential for the formation of a highly condensed subtelomeric chromatin body called the knob. Furthermore, Sgo2 maintains the expression levels of subtelomeric genes and the timing of DNA replication at subtelomeric late origins.
The telomere, a specialized chromatin structure at the ends of chromosomes, plays important roles in genome integrity, and the adjacent chromosomal region, the subtelomere, has recently become a focus of attention in the medical field. Subtelomere deletion syndrome (STD) is a congenital disease arising from deletion or rearrangement in the subtelomeric region. STD patients exhibit mental retardation and multiple congenital anomalies, which most likely develop from the haploinsufficiency of subtelomeric genes (de Vries et al., 2003). Furthermore, facioscapulohumeral muscular dystrophy is caused by an increase in the expression of the DUX4 gene, located at the subtelomere, which is affected by telomere position effects (Stadler et al., 2013). Therefore, maintenance of subtelomeric gene expression appears to be critical for human health.
Although substantial knowledge of telomeres has accumulated, the regulation and function of subtelomeres remain a long-term mystery: research on subtelomeres has been progressing much more slowly than research on other chromosomal regions due to technical difficulties resulting from the structural nature of subtelomeric homologous sequences, which are very long and present in large numbers of copies. In particular, how the condensed and silenced chromatin structure at subtelomeres is formed and how replication timing at subtelomeres is regulated remain outstanding issues.
Composition of subtelomeric DNATelomeres are composed of DNA with repeated sequence ([TTAGGG]n in mammals) and its associated proteins. In contrast, subtelomeres do not possess telomere repeat sequences; however, they generally contain multiple segments that share high similarity with each other in a specific organism, in addition to various genes. In budding yeast, Saccharomyces cerevisiae, subtelomeres span approximately 25 kb and include X and Y’ elements with highly variable lengths. The X element contains a highly homologous X core region (475 bp) and an autonomously replicating sequence; the Y’ element contains an RNA helicase gene (Louis, 1995). Internal repeat sequences exist in some subtelomeres at the telomere-distal region adjacent to the X’ and Y elements. The length of the internal repeats is also variable; they sometimes contain genes encoding PAU (seripauperin multigene family), MAL (multigene complex for maltose fermentation), MEL (α-galactosidase) or ERR (enolase) proteins (Louis, 1995) (Fig. 1A). In humans, subtelomeres possess a mosaic of multiple common sequences (more than 40 types in total) containing various genes. The length of the homologous regions of human subtelomeres varies from less than 20 kb to more than 200 kb (Linardopoulou et al., 2005; Riethman et al., 2005; Stong et al., 2014).
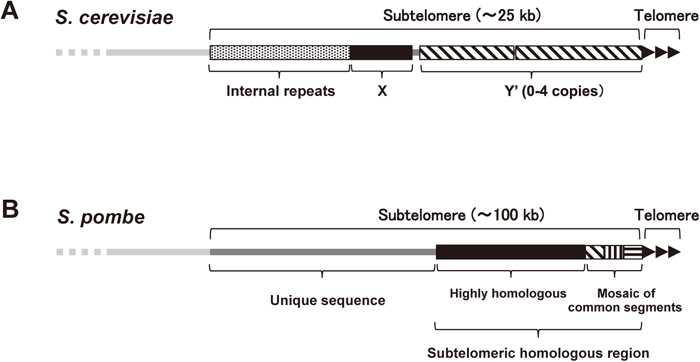
Composition of subtelomeres. (A) Representative structure of the subtelomeres in S. cerevisiae. (B) Structure of the subtelomeres of chromosomes 1 and 2 in S. pombe. See main text for details.
The fission yeast, Schizosaccharomyces pombe, possesses three chromosomes. Although genome sequencing is not yet complete, the subtelomeres of S. pombe also contain highly homologous sequences spanning 40–60 kb in chromosomes 1 and 2 that harbor the RecQ-type helicase genes (tlh1/2+) (http://www.pombase.org/status/sequencing-status). The telomere-proximal regions are composed of a mosaic of common segments exhibiting variations like those in humans (Sugawara, 1988). The telomere-distal regions of approximately 50 kb do not contain sequences that are homologous between subtelomeres. However, they share a common feature of chromatin structure: the levels of histone modifications, such as methylation and acetylation, are very low compared with the adjacent euchromatin and subtelomeric heterochromatin regions (see below) (Buchanan et al., 2009) (Fig. 1B and 2). The number of subtelomeres varies from four to six per S. pombe haploid genome, depending on the presence or absence of a short subtelomereric homologous sequence (approximately 15 kb) and a chromosome 3-specific sequence (1.1 kb) between the telomeres and the rDNA repeats at the ends of chromosome 3 (Ohno et al., 2016).

Schematic illustration of the chromatin structure of S. pombe subtelomeres. Heterochromatin containing highly methylated H3K9 and Swi6 is formed around the subtelomeric homologous region (approximately 50 kb). Sgo2 associates with the whole subtelomere (approximately 100 kb), and is required for the formation of a knob (approximately 50 kb) next to the heterochromatin, where various histone modifications are maintained at low levels. A euchromatin region, where gene transcription is active, is adjacent to the subtelomere.
Subtelomeres generally include heterochromatin, where histone H3 methylated at K9 (H3K9me) is highly enriched and heterochromatin protein 1 (HP1) homologs are associated (Elgin and Grewal, 2003). At least two factors can independently establish subtelomeric heterochromatin in S. pombe. One is the telomere-binding protein complex called shelterin; the other is the RNAi machinery, which acts at the centromere-homologous (cenH) sequence (the template of siRNA) within the tlh1/2+ genes in the subtelomeres. After a methyltransferase, Clr4, methylates H3K9 and an HP1 homolog, Swi6, is recruited to the telomere-proximal sites, the heterochromatin spreads out around the subtelomeric homologous regions, presumably via the association of Swi6 with itself and Clr4 (Hall et al., 2002; Cam et al., 2005; Kanoh et al., 2005; Wang et al., 2016) (Fig. 3A).

Formation of subtelomeric heterochromatin. (A) In S. pombe, the shelterin proteins (Taz1 or Ccq1) associated with the telomeres and with the telomere-proximal region of the subtelomere and RNAi machinery acting at the cenH sequence independently recruit Clr4 to methylate H3K9. H3K9me recruits Swi6 (HP1), and then the heterochromatin spreads out to the chromosomal inner region. (B) In S. cerevisiae, the telomere-binding protein Rap1 recruits Sir3-Sir4, which in turn recruits Sir2 to deacetylate histones H3 and H4. Sir3-Sir4 then associates with the deacetylated histones and again recruits Sir2 to expand the heterochromatin region.
In contrast, S. cerevisiae does not possess H3K9me or HP1 homologs; instead, the telomere-binding protein Rap1 associates with Sir3-Sir4 (silencing factors), which then recruits Sir2, a histone deacetylase, to deacetylate histones H3 and H4. After this, Sir3-Sir4 is again recruited to the deacetylated chromatin and again recruits Sir2, to expand the heterochromatin (Huang, 2002) (Fig. 3B).
Heterochromatin generally brings about a transcriptionally silent state. Thus, the expression of genes located within subtelomeric heterochromatin is repressed; however, the physiological functions of these gene products remain largely unknown. Moreover, the roles of subtelomeric heterochromatin itself have not yet been clarified.
Sgo2 is localized at subtelomeres during interphaseShugoshin is a well conserved centromere protein and plays crucial roles in centromeric cohesion and/or proper chromosome segregation in mitosis and meiosis (Kerrebrock et al., 1995; Kitajima et al., 2004; Marston et al., 2004; Rabitsch et al., 2004; Salic et al., 2004; Kawashima et al., 2007; Vanoosthuyse et al., 2007). Schizosaccharomyces pombe possesses two shugoshin paralogs, Sgo1 and Sgo2. Sgo1 is expressed only during meiosis and plays a critical role in centromeric cohesion of sister chromatids in meiosis I (Kitajima et al., 2004). In contrast, Sgo2 is continuously expressed and contributes to precise chromosome segregation during M phase (Kawashima et al., 2007). Intriguingly, it was reported that Sgo2 changes its localization from the centromeres to the vicinity of the telomeres upon entry into interphase (Kawashima et al., 2010); however, detailed information about Sgo2 localization and the physiological roles of Sgo2 near telomeres remained unknown. In the following sections, I describe our recent discovery of unexpected roles of Sgo2 at the subtelomeres in S. pombe (Tashiro et al., 2016).
Chromatin immunoprecipitation (ChIP)–chip analyses revealed that Sgo2 is enriched not only at the centromeres but also throughout the subtelomeres of both chromosomes 1 and 2, extending more than 100 kb from each telomere (Fig. 2). However, Sgo2 was barely detectable near the telomeres of chromosome 3 in a strain that possesses no subtelomeric sequence in chromosome 3. ChIP analyses using synchronous cell culture showed that Sgo2 associates with the subtelomeres preferentially during interphase, with a peak in the middle of G2 phase.
Phosphorylation of histone H2A is important for Sgo2 localizationWhat is the requirement for localization of Sgo2 at the subtelomeres? Sgo2 remains attached to the subtelomeres in cells lacking telomeric DNA, indicating that the telomeric structure is dispensable for the association of Sgo2 with the subtelomeres. In contrast, deletion of bub1+, which encodes the sole protein kinase that phosphorylates serine 121 of histone H2A (H2A-S121) for the activation of the spindle assembly checkpoint (Kawashima et al., 2010), abolishes the subtelomeric localization of Sgo2 almost completely. Substituting H2A-S121 with a non-phosphorylatable alanine (h2a-S121A) or the bub1-KD (kinase-dead) mutation results in almost the same level of removal of Sgo2 from the subtelomeres as observed in bub1∆. Therefore, the phosphorylation of H2A-S121 by Bub1 is important for the association of Sgo2 with subtelomeres.
Sgo2 is crucial for the formation of a highly condensed chromatin body, the knobWhat are the functions of Sgo2 at the subtelomeres in interphase? Knobs are highly condensed chromatin bodies (more condensed than heterochromatin) in S. pombe that are intensely stained with the DNA-labeling dye DAPI (4’,6-diamidino-2-phenylindole). They are located in the vicinity of the telomeres, adjacent to the subtelomeric heterochromatin (marked by H3K9me), specifically during interphase (Matsuda et al., 2015). Interestingly, the subnuclear localization of Sgo2 always overlaps with the knobs, which are completely lost in sgo2Δ cells. Consistent with this, bub1Δ and h2a-S121A mutant cells, in which Sgo2 is dissociated from the subtelomeres, also completely lack knobs. Thus, the association of Sgo2 with subtelomeres plays a crucial role in knob formation.
Sgo2 represses transcription of subtelomeric genesWhat is the consequence of Sgo2-mediated knob formation? Condensed chromatin is generally expected to be inhibitory to transcription (Politz et al., 2013). Genome-wide microarray analyses showed that the transcript levels of genes at subtelomeres are increased in sgo2Δ cells, whereas gene expression is not strongly affected by sgo2Δ in other chromosomal regions including centromeres, where Sgo2 is highly enriched in M phase. Notably, the locations of the genes affected by sgo2Δ correlate with the subtelomeric distribution of Sgo2. Transcription of subtelomeric genes is also derepressed in the bub1Δ and h2a-S121A mutants, and RNA polymerase II (RNAPII) occupancy at subtelomeric gene loci is considerably higher in the sgo2∆, bub1∆ and h2a-S121A mutants than in the wild-type strain. Furthermore, the levels of histone modifications associated with transcriptionally active chromatin, such as dimethylation of lysine 4 and trimethylation of lysine 36 in histone H3 (Noma et al., 2001; Kizer et al., 2005; Morris et al., 2005), are increased at subtelomeric gene loci in sgo2∆, bub1∆ and h2a-S121A cells. Collectively, these studies indicate that Sgo2 maintains gene expression at the transcriptional level, accompanied by histone modifications, specifically at the subtelomeres.
Sgo2 forms a new type of repressive chromatin domainAs described above, heterochromatin generally inhibits the expression of nearby genes. Do Sgo2 and subtelomeric heterochromatin regulate gene expression independently of each other? Monitoring the expression of an exogenous marker gene inserted at a telomere-proximal site within a subtelomere demonstrated that Swi6-heterochromatin is dominant to repress gene expression in the telomere-proximal region. In contrast, there was a marked increase in the expression of the marker gene when it was placed at a telomere-distal site within a subtelomere in sgo2∆ and bub1∆ cells, whereas only a small increase was observed in swi6∆ cells, indicating that the subtelomeric localization of Sgo2 is required for this repressive effect (Tashiro et al., 2016). These results indicate that the association of Sgo2 with subtelomeres forms a new type of repressive chromatin domain that is distinct from H3K9me-Swi6-mediated heterochromatin.
Sgo2 maintains replication timing at subtelomeresDoes the subtelomeric localization of Sgo2 have other effects on cellular functions? Schizosaccharomyces pombe subtelomeres contain many DNA replication origins that fire during late S phase (Hayashi et al., 2007). The replication timing of some subtelomeric late origins is regulated by the telomere-binding proteins Taz1 and Rif1 (Hayano et al., 2012; Tazumi et al., 2012) (see Hisao Masai et al.’s review in this issue); however, the overall regulatory mechanism of subtelomeric replication timing remains elusive. Intriguingly, the clusters of late origins at the subtelomeres largely overlap with Sgo2 localization. Indeed, in sgo2Δ, bub1-KD and h2a-S121A cells, the subtelomeric late origins replicate earlier than in wild-type cells, whereas the replication timing in other chromosomal regions does not change (Tashiro et al., 2016). Thus, the association of Sgo2 with subtelomeres has a repressive effect on replication to maintain proper replication timing at the subtelomeric late origins.
Sgo2 regulates loading of Sld3 onto subtelomeric replication originsHow does Sgo2 regulate replication timing? A replication factor, Sld3, is recruited to origins during the earliest step of the activation of pre-replicative complex upon S phase entry (Yabuuchi et al., 2006). Sld3 is enriched at the early origins but not at the late origins in wild-type cells arrested immediately before the stage of replication initiation. In contrast, in sgo2Δ cells, recruitment of Sld3 is accelerated at the subtelomeric late origins, but not at the internal late origins. Therefore, Sgo2 represses the initiation step of replication by limiting Sld3 loading at the subtelomeric late origins, which is required for proper replication timing.
Conclusion and future prospectsSgo2 is localized at the centromeres and contributes to precise chromosome segregation in M phase. On the other hand, during interphase, Sgo2 is recruited to the subtelomeres and plays important roles in the formation of knobs, in the repression of gene transcription, and in the repression of replication to ensure proper gene expression and replication timing at the subtelomeres. Thus, Sgo2 is a multifunctional protein shuttling between centromeres and subtelomeres during the cell cycle (Tashiro et al., 2016) (Fig. 4).

Sgo2 plays various roles at the centromeres and subtelomeres in S. pombe. Sgo2 is localized at the centromeres in M phase for precise chromosome segregation. On the other hand, during interphase, Sgo2 changes its localization to the subtelomeres and regulates knob formation, the expression of subtelomeric genes, and the replication timing of subtelomeric late origins.
Many questions have been raised from the study of Sgo2. First, how is Sgo2 targeted to the subtelomeres during interphase? Phosphorylation of H2A-S121 is observed outside the subtelomeres in interphase (Tashiro et al., 2016), indicating that H2A-S121 phosphorylation alone is not sufficient and that another factor is required to confine Sgo2 to the subtelomeres. Because Sgo2 dynamically changes its location during the cell cycle, some cell-cycle regulators and/or modifications of Sgo2 (such as phosphorylation) may be involved in Sgo2 recruitment to the subtelomeres.
Second, how does Sgo2 regulate knob formation, gene expression and replication timing? Is it related to the functions of Sgo2 at centromeres? One possibility is that Sgo2 regulates subtelomeric gene expression and replication timing through the knobs. The highly condensed knob structure may inhibit the recruitment of RNAPII, histone modification enzymes and replication factors. The other possibility is that Sgo2 acts at the subtelomeres independently of the knobs. As described in Masai et al.’s review, the telomere-binding protein Rif1 regulates the replication timing of late origins by interacting with PP1 phosphatase to counteract the action of Dbf4-dependent kinase (DDK) at the initiation step of replication (Davé et al., 2014; Hiraga et al., 2014; Mattarocci et al., 2014). Interestingly, Sgo1 forms a complex with PP2A phosphatase to protect cohesins in S. pombe, S. cerevisiae and mammals (Kitajima et al., 2006; Riedel et al., 2006; Tang et al., 2006; Ishiguro et al., 2010), and human Sgo2 also interacts with PP2A for the protection of cohesion (Tanno et al., 2010). Thus, S. pombe Sgo2 may also interact with a protein phosphatase to counteract the activity of DDK and thereby regulate Sld3 loading.
Third, how is the highly condensed knob structure formed? Intriguingly, knobs are not formed at the centromeres (Matsuda et al., 2015), despite the high accumulation of Sgo2 at that location in M phase, suggesting that Sgo2 by itself is not sufficient and requires other factors to form knobs. The mechanism underlying the subtelomere-specific formation of knobs remains unclear; however, one possibility is that a unique chromatin structure with low levels of acetylated and methylated histones (Buchanan et al., 2009) contributes to knob formation. The low level of H3K9 methylation in the telomere-distal region of the subtelomeres (adjacent to the subtelomeric heterochromatin region) is in sharp contrast to the high level of H3K9 methylation in the Sgo2-associated outer regions of the centromeres (Kawashima et al., 2007; Buchanan et al., 2009). In fact, knobs are observed preferentially in the telomere-distal regions of the subtelomeres (Matsuda et al., 2015) (Fig. 2).
Fourth, are the shugoshin family proteins in other organisms also involved in subtelomere functions? Subtelomeric localization of shugoshin in other organisms has not yet been reported. In humans, however, maintenance of the expression levels of subtelomeric genes is important for health. Thus, one can speculate that there exist some regulatory mechanisms, possibly involving shugoshin, for the local organization of a repressive chromatin domain at subtelomeres in other eukaryotes, as in S. pombe.