2018 Volume 93 Issue 1 Pages 31-35
2018 Volume 93 Issue 1 Pages 31-35
Microsatellite markers were developed for the endangered orchid Calanthe izu-insularis (Orchidaceae). This species is unique to the Izu Islands in Japan. Unfortunately, its population size has decreased because of excessive collection for horticultural purposes. In addition, although natural hybridization between C. izu-insularis and C. discolor var. discolor has been reported, morphological differences between C. izu-insularis and the hybridized individuals remain unclear. Using next-generation sequencing, 11 polymorphic microsatellite markers were developed. All developed markers could amplify C. aristulifera and nine markers could amplify C. d. var. discolor, two other orchid species that are also endangered in Japan. The number of alleles and expected heterozygosity at each locus were 1–6 (mean, 2.35) and 0.00–0.79 (mean, 0.30), respectively. These microsatellite markers will help conservation geneticists in their investigation of the proportion of pure C. izu-insularis individuals in the Izu Islands.
Calanthe izu-insularis (Satomi) Ohwi et Satomi (Orchidaceae) is an endemic perennial plant species in the Izu Islands, Japan (Cribb and Bailes, 2001). This species is now classified as endangered according to the Red List of Japan (http://www.env.go.jp/press/files/jp/105449.pdf). The main factor for the decline of C. izu-insularis is excessive collection for horticulture because of the rare, beautiful and fragrant nature of this orchid (Ministry of the Environment, Government of Japan, 2015). Recently, the collection of wild C. izu-insularis has been prohibited in Mikura and Kodzu Islands; however, the risks of illegal collection persist. Thus, ubiquitous genotyping is essential for the conservation of this plant (Shiga et al., 2013; Isagi and Kaneko, 2014). Ubiquitous genotyping data would be based on all remaining individuals and would be useful for assisting in the prevention of their illegal collection (Shiga et al., 2013).
Based on morphological data, C. izu-insularis is closely related to two other species, C. aristulifera and C. discolor var. discolor, which are widely distributed throughout the islands of Japan (Cribb and Bailes, 2001). Natural hybridization has also been reported between C. izu-insularis and C. d. var. discolor (Takahashi, 1972; Cribb and Bailes, 2001). These two species sympatrically grow in the Izu Islands (Cribb and Bailes, 2001; Suetsugu et al., 2016). In addition, they are pollinated by the same bee species, Lasioglossum occidens (Suetsugu et al., 2016, 2017). Although the morphological difference between C. izu-insularis and the hybridized individuals is unclear, the application of molecular techniques should make it possible to distinguish them.
In this study, we developed 11 microsatellite markers for C. izu-insularis using next-generation sequencing and examined the availability of these markers for two related species of Calanthe, namely C. aristulifera and C. d. var. discolor (Table 1).
| Locus | Primer sequences (5’-3’) | Repeat motif | Fluorescent labela | Ta (℃) | Size range (bp) | Total number of alleles in all species | Species used for primer design | GenBank Accession No. |
|---|---|---|---|---|---|---|---|---|
| Caiz_005 | F: GTGCACAACACATGAAGTCTG | (CT)9 | FAM | 57 | 422–428 | 3 | C. izu-insularis | LC271144 |
| R: TCTTTAGGTTATGTAACACCAGGG | ||||||||
| Caiz_010 | F: CCATCCGTCAAACCAACGG | (TC)10 | VIC | 57 | 332–345 | 8 | C. izu-insularis | LC271145 |
| R: AAATTCGCGGCGGACAAAG | ||||||||
| Caiz_020 | F: AGCCTAAGAGTAAGGTTGAAATTGG | (TG)10 | NED | 57 | 226–250 | 11 | C. izu-insularis | LC271146 |
| R: TGGCATTTGACAAGGGCTATTC | ||||||||
| Caiz_024 | F: AACCCGACGCCCAGAATAG | (AC)8 | FAM | 57 | 255–257 | 2 | C. izu-insularis | LC271147 |
| R: TGATAACCACCAAGCAAGGAC | ||||||||
| Caiz_036 | F: CATCTGGTGCTATCACGCC | (GA)9 | FAM | 57 | 299–339 | 3 | C. izu-insularis | LC271148 |
| R: TGCATAAGGTCATCAATCCTTTAC | ||||||||
| Caiz_041 | F: TCAGGCTCTAACTTGTTGGG | (AAC)8 | NED | 57 | 282–296 | 4 | C. izu-insularis | LC271149 |
| R: TCTGCCTCAAATTCATGGACTG | ||||||||
| Caiz_045 | F: CTCAACAGCATTGGCCTCG | (TGA)6 | NED | 57 | 352–373 | 6 | C. izu-insularis | LC271150 |
| R: AGAAGCAACCCGAAACATCC | ||||||||
| Caiz_047 | F: GAGCCTGGTCTTGTGCATC | (AAG)7 | VIC | 57 | 266–270 | 3 | C. izu-insularis | LC271151 |
| R: TGCCTAGGATTTCTTTGGCTG | ||||||||
| Caiz_068 | F: TGCTAAGTTAAACCCTGTGTGC | (AAT)5 | NED | 57 | 167–170 | 2 | C. izu-insularis | LC271152 |
| R: ATTCAAACTCGAGGGCTTGG | ||||||||
| Caiz_071 | F: GGGACTCGCACATCCCTAG | (CCG)9 | VIC | 57 | 127–145 | 7 | C. discolor var. discolor | LC271153 |
| R: GTAAGGCGTCGCTGCCCTCGC | ||||||||
| Caiz_083 | F: CGGCTGAGGACCCCGTCTT | (CGG)6 | FAM | 57 | 214–223 | 4 | C. discolor var. discolor | LC271154 |
| R: TCCGTTCGACATTATGGCCG |
Note: Ta annealing temperature.
To develop microsatellite markers for C. izu-insularis and related species, we constructed DNA libraries from C. izu-insularis, C. d. var. discolor and C. d. var. kanashiroi. Fresh leaf samples of one individual of C. izu-insularis (collected from Mikura Island, Tokyo prefecture, Japan), two individuals of C. d. var. discolor (collected from Mikura Island, Tokyo prefecture and Tokunoshima Island, Kagoshima Prefecture) and one individual of C. d. var. kanashiroi (collected from Okinawa Island, Okinawa Prefecture) were used for DNA extraction. Genomic DNA of all species was extracted using the DNeasy Plant Mini Kit (Qiagen), nebulized at 0.24 MPa for 1 min and purified using the MinElute PCR Purification Kit (Qiagen).
The purified DNA fragments of C. izu-insularis underwent end repair and A-tailing using the Rapid Library Prep Kit (Roche) and were ligated to the Rapid Library Adapter (Roche) with RL Ligase (Roche). Suitably sized DNA fragments were selected by removing short fragments using AMPure XP beads (Agilent). Fragments were then mixed with capture beads and amplified using emulsion polymerase chain reaction (emPCR) with the GS Junior Titanium emPCR kit (Roche). After emPCR, beads were collected and those that captured the DNA library were enriched before annealing with sequencing primers. Amplified fragments were sequenced using the GS Junior Bench Top system (Roche). Consequently, a total yield of 35,982 reads was achieved.
Genomic DNA from two individuals of C. d. var. discolor and one individual of C. d. var. kanashiroi was digested with MspI and PstI (New England Biolabs) at 37 ℃ for 2 h. Adapters were ligated at 22 ℃ overnight in a 40-μl solution containing 20 μl of digested DNA, 2 μl of 10× NEB buffer 4, 0.02 μM adapter 1, 3 μM adapter 2, 1 mM ATP and 0.5 μl of T4 DNA ligase (Enzymatics). The DNA was then purified with the QIAquick PCR purification kit (Qiagen), and 7.5 μl of purified DNA was used for PCR amplification in a 25-μl solution containing 12.5 μl of 2× Multiplex PCR Master Mix (Qiagen), 0.2 μM Illumina forward primer and 0.2 μM Illumina reverse primer. Thermal cycling was initiated with a 95 ℃ step for 15 min, followed by 27 cycles of 94 ℃ for 30 s, 62 ℃ for 1.5 min and 72 ℃ for 1.5 min and a final extension at 72 ℃ for 10 min. PCR products were purified again with the QIAquick PCR purification kit. DNA fragments of 500–600 bp were extracted using Pippin Prep (Sage Science). The amplification and extraction was checked using MultiNA (Shimadzu), and the fragments were sequenced using the Illumina Miseq v2 (Illumina). The total numbers of reads obtained from C. d. var. discolor and C. d. var. kanashiroi were 4,003 and 1,228, respectively.
The reads were screened using PRIMER3 software (Rozen and Skaletsky, 2000) embedded in msatcommander version 0.8.2 (Faircloth, 2008) to identify dinucleotide and trinucleotide loci of 5–18 repeats. A total of 90 primer pairs were designed, comprising 68, 18 and 4 primer pairs from C. izu-insularis, C. d. var. discolor and C. d. var. kanashiroi reads, respectively.
Primer checkAmplification by and specificity of the primer pairs were tested using four C. izu-insularis and four C. d. var. discolor individuals. For all confirmed microsatellite loci in C. izu-insularis, locus-specific forward primers were synthesized with one of the following M13 sequence tails, 5’–CACGACGTTGTAAAACGAC–3’, 5’–CTATAGGGCACGCGTGGT–3’, 5’–TGTGGAATTGTGAGCGG–3’ (Boutin-Ganache et al., 2001), added to their 5’ ends (Table 1). Each PCR amplification was performed in a final volume of 5 μl containing approximately 16 ng of template DNA, 2.5 μl of 2× Multiplex PCR Master Mix (Qiagen), 0.01 μM forward primer, 0.2 μM reverse primer and 0.1 μM M13 (fluorescent-labeled) primer. The PCR thermal profile was as follows: initial denaturation at 95 ℃ for 15 min, followed by 30–35 cycles of 94 ℃ for 30 s, 57 ℃ for 1.5 min and 72 ℃ for 1 min and a final extension at 60 ℃ for 30 min. PCR products were detected on the ABI Prism 3130 Genetic Analyzer (Applied Biosystems). Fragment lengths were calculated using Gene Mapper (Applied Biosystems).
Polymorphism of microsatellite markersFor population genetic analysis, DNA was extracted from 78, 3 and 31 individuals of C. izu-insularis, C. aristulifera and C. d. var. discolor using the CTAB method (Milligan, 1992). Leaf samples of C. izu-insularis and C. aristulifera were collected from Mikura Island, and those of C. d. var. discolor were collected from two populations located in Ibaraki and Hiroshima Prefectures. PCR conditions were the same as used for the above primer check. The following three summary statistics were calculated using GenAlEx version 6.4 (Peakall and Smouse, 2006) and FSTAT version 2.9.3 (Goudet, 1995): number of alleles (A), observed heterozygosity (HO) and expected heterozygosity (HE). Tests for deviation from Hardy–Weinberg equilibrium (HWE) and for linkage disequilibrium (LD) between loci were performed using FSTAT version 2.9.3. The combined first-parent non-exclusion probabilities for an identity were calculated using Cervus 3.0 for each species (Marshall et al., 1998; Kalinowski et al., 2007). These probabilities were calculated using 11 markers in C. izu-insularis and C. aristulifera and nine markers in C. d. var. discolor.
To evaluate genetic differentiation among C. izu-insularis, C. aristulifera and C. d. var. discolor, we calculated pairwise codominant genotypic distances (Smouse and Peakall, 1999) among each species. We also performed principal coordinates analysis (PCoA) using GenAlEx version 6.41 (Peakall and Smouse, 2006).
Of the 90 primer pairs tested, 11 amplified all four C. izu-insularis and four C. d. var. discolor individuals. The lengths of the amplified fragments were unambiguously scored. Their size range for each primer pair is shown in Table 1. Nine and two primer pairs were designed from the library of C. izu-insularis and C. d. var. discolor, respectively. Four primer pairs designed from the library of C. d.var. kanashiroi were unusable for amplification.
All primers successfully amplified all of the samples of C. izu-insularis and C. aristulifera; however, two primer pairs, Caiz_005 and Caiz_024, could not amplify 27 and 23 samples, respectively, of a total of 31 C. d. var. discolor samples (Table 2).
| Locus | Calanthe izu-insularis | Calanthe aristulifera | Calanthe discolor var. discolor | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mikura Island (N = 78) | Mikura Island (N = 3) | Ibaraki (N = 14) | Hiroshima (N = 17) | |||||||||
| A | HO | HE | A | HO | HE | A | HO | HE | A | HO | HE | |
| Caiz_005 | 2 | 0.35 | 0.39 | 1 | 0.00 | 0.00 | – | – | – | – | – | – |
| Caiz_010 | 2 | 0.27 | 0.37 | 3 | 1.00 | 0.61 | 4 | 0.64 | 0.59 | 6 | 1.00 | 0.79 |
| Caiz_020 | 4 | 0.45 | 0.42 | 3 | 1.00 | 0.61 | 6 | 0.50 | 0.79 | 5 | 0.76 | 0.70 |
| Caiz_024 | 2 | 0.13 | 0.14 | 1 | 0.00 | 0.00 | – | – | – | – | – | – |
| Caiz_036 | 2 | 0.03 | 0.03 | 1 | 0.00 | 0.00 | 2 | 0.07 | 0.07 | 1 | 0.00 | 0.00 |
| Caiz_041 | 2 | 0.01 | 0.01 | 2 | 0.33 | 0.28 | 1 | 0.00 | 0.00 | 1 | 0.00 | 0.00 |
| Caiz_045 | 2 | 0.06 | 0.06 | 3 | 1.00 | 0.61 | 2 | 0.43 | 0.35 | 2 | 0.12 | 0.11 |
| Caiz_047 | 1 | 0.00 | 0.00 | 1 | 0.00 | 0.00 | 2 | 0.14 | 0.53 | 2 | 0.24 | 0.21 |
| Caiz_068 | 1 | 0.00 | 0.00 | 1 | 0.00 | 0.00 | 1 | 0.00 | 0.00 | 1 | 0.00 | 0.00 |
| Caiz_071 | 2 | 0.40 | 0.45 | 3 | 0.33 | 0.61 | 4 | 0.36 | 0.48 | 4 | 0.82 | 0.70 |
| Caiz_083 | 3 | 0.46 | 0.49 | 3 | 0.67 | 0.50 | 3 | 0.86 | 0.55 | 2 | 0.18 | 0.41 |
| Average | 2.09 | 0.20 | 0.22 | 2.00 | 0.39 | 0.29 | 2.78 | 0.33 | 0.37 | 2.67 | 0.35 | 0.32 |
Note: N = sample size; A = number of alleles; HO = observed heterozygosity; HE = expected heterozygosity.
The number of alleles per locus ranged from 1 to 6 (mean, 2.35) (Table 2). The observed (HO) and expected (HE) heterozygosities per locus ranged from 0.00 to 1.00 (mean, 0.32) and from 0.00 to 0.79 (mean, 0.30), respectively (Table 2). The total number of alleles in all three species ranged from 2 to 11 (mean, 4.82) (Table 1). All 11 loci showed no evidence of significant deviation from HWE (P > 0.05). Significant LD was observed in the Ibaraki and Hiroshima populations of C. d. var. discolor for two pairs of loci (Caiz_010 and Caiz_020, Caiz_020 and Caiz_071; P < 0.05). In the Hiroshima populations of C. d. var. discolor, there was also significant LD for an additional pair of loci (Caiz_010 and Caiz_071; P < 0.05). The combined non-exclusion probabilities for an identity in C. izu-insularis, C. aristulifera and C. d. var. discolor were 0.0069, 0.0004 and 0.0001, respectively. According to PCoA based on codominant genotypic distances, 64.49% and 5.17% of the variation was described with the first and second axes, respectively (Fig. 1). Although individuals between C. izu-insularis and C. aristulifera were not clearly divided, C. izu-insularis and C. d. var. discolor individuals were plotted as independent groups. This may be attributed to the small number of samples of C. aristulifera.
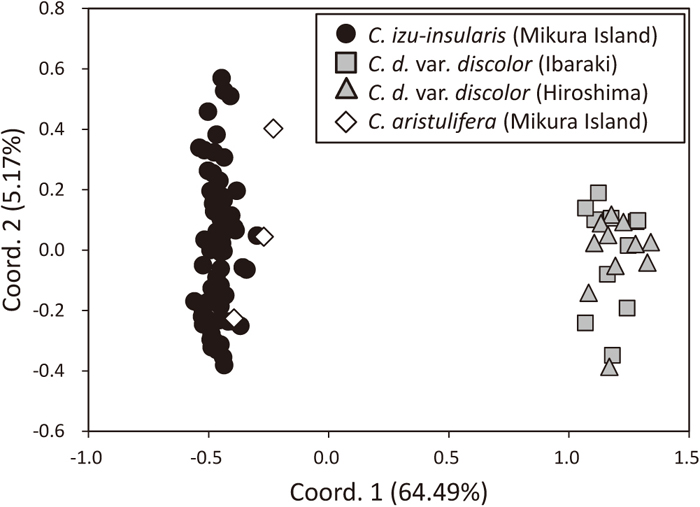
Principal coordinates analysis plot of individuals based on codominant genotypic distances (Smouse and Peakall, 1999). Axis 1 explains 64.49% of the variance and axis 2 explains 5.17% of the variance.
Performing population genetic analysis using these microsatellite markers may contribute to the conservation of C. izu-insularis. Considering that the main factor for the decline of C. izu-insularis is excessive collection for horticulture, the use of these markers to identify genotypes of C. izu-insularis could prevent their illegal collection (Shiga et al., 2013). Furthermore, the assessment of hybridization between C. izu-insularis and C. d. var. discolor should enable us to determine the proportion of pure C. izu-insularis individuals.
These markers may be capable of identifying other species of Calanthe, such as C. aristulifera. As described in the Red List of Japan, several species of Calanthe are now classified as near threatened, endangered or critically endangered (http://www.env.go.jp/press/files/jp/105449.pdf). Therefore, population genetic studies of the species of Calanthe, such as those evaluating genetic diversity and structure within and between populations, are essential for constructing successful conservation programmes.
We are grateful to Mr. Kuniaki Ohtake, Dr. Tomohisa Yukawa and the Hiroshima Botanical Garden for providing C. d. var. discolor samples. We are also grateful to Mr. Masayuki Hino and Mr. Toshiaki Kurimoto for helping us to collect samples of C. aristulifera and C. izu-insularis from Mikura Island. This work was supported by JSPS KAKENHI Grant Number 15H04414 and the Environment Research and Technology Development Fund (4-1605) of the Ministry of the Environment, Japan.