2018 Volume 93 Issue 2 Pages 75-81
2018 Volume 93 Issue 2 Pages 75-81
Paired box (Pax) 6, a member of the Pax family of transcription factors, contains two DNA-binding domains, called the paired domain (PD) and the homeodomain (HD), and plays pivotal roles in development of structures such as the eye, central nervous system and pancreas. Pax6 is a major developmental switching molecule because, for example, ectopic expression of the Pax6 gene can induce ectopic whole eye development. Intensive research has been devoted to elucidating the molecular mechanism(s) involved in the function(s) of Pax6, but many issues remain unexplained. One of the important issues is to identify the nuclear localization signal (NLS) in the PD of Pax6, which is predicted to have a stronger NLS activity than that in the HD. We produced expression plasmid constructs that encode the chick Pax6 protein modified to delete the entire PD except for fragments containing putative NLS sequences, and electroporated them in ovo into the developing chick midbrain to define the NLS of the PD. The results show that the NLS in the PD of chick Pax6 consists of an unusually long sequence of 36 amino acid residues. Within this long NLS motif, the central 18 amino acids comprising two consecutive nine-residue segments showed highest NLS activity; this central area corresponds to the C-terminal half of the third α–helix of the PAI subdomain and the subsequent 11 amino acids of a 16-residue linker between PAI and the adjacent RED subdomain. This information helps to elucidate the molecular mechanism by which Pax6 plays a pivotal role during ontogeny.
Paired box (Pax) 6 belongs to the Pax family of transcription factors, which are highly conserved and are involved in regulating the morphogenesis of many tissues and organs through ontogeny, playing pivotal roles in, for example, development of the eye, central nervous system and pancreas. Pax6 is so potent that ectopic expression of the mouse and Xenopus Pax6 genes can induce ectopic eye development in the fruit fly (Halder et al., 1995; Gehring and Ikeo, 1999). According to Cvekl and Callaerts (2017), the Pax6 gene is one of a small group that encodes master regulatory proteins that switch molecular networks, initiating the development of specific cell types. Elucidating the functional role of Pax6 during development is an essential step for the phylogenetic comparison of the developmental mechanism(s) in which it is involved.
Pax6, like most other members of the Pax family, consists of two DNA-binding domains, the paired domain (PD) and the homeodomain (HD). Compared to our accumulated knowledge about the structure and function of the HD, the PD is not well understood and needs further study. One such issue is to clarify the amino acid residues responsible for the function of the nuclear localization signal (NLS) in the PD (PD-NLS). Regarding the PD-NLS, Carrière et al. (1993, 1995) reported that deletion of exon 5 of quail (Coturnix coturnix japonica) Pax6 (Pax-QNR) significantly decreased the localization of the resulting Pax6 isoform in the nucleus. They predicted a very strong NLS in the amino acid sequence encoded by exon 5 of the Pax-QNR, which would be more active than the NLS that spans the N-terminus of the HD (HD-NLS, also called HNLS5 [LKRKLQR, residues L206 – R212]) (Carrière et al., 1995). Therefore, the function of Pax6 may heavily depend on the NLS in the PD domain. In this sense, it is crucial to localize the amino acid sequence comprising the NLS of the PD. Identifying that sequence would help us not only to infer the functional evolution of the PD domain, which has had a crucial role ever since the Tc1/mariner transposon was captured and domesticated in the ancient Pax family (Paixão-Côrtes et al., 2015), but also to design expression constructs, such as those harboring the nucleotide sequence encoding the NLS of the PD without any DNA-binding activity, to dissect the molecular function(s) of the PD in ontogeny.
According to Carrière et al. (1995), whose experiments were carried out in vitro using COS and quail embryonic cells, it is possible that all amino acid residues encoded by exon 5 of quail Pax6 can act as a NLS. If that is the case, an exceptionally long region of amino acid sequence would comprise the NLS, since 72 amino acid residues are encoded by exon 5 of quail Pax6 (gene ID: X70475.1; protein ID CAA49899.1), which is homologous to chick Pax6 exon 6 (Fig. 1A). NLSs are usually only seven to 10 residues in length: for example, the c-myc NLS contains only nine amino acid residues (PAAKRVKLD, Dang and Lee, 1988) and the p53 NLS contains only seven residues (PQPKKKP, Liang and Clarke, 1999). To clarify this issue, we now report our functional analysis in ovo to survey short amino acid sequences contributing to the PD-NLS in chick Pax6.
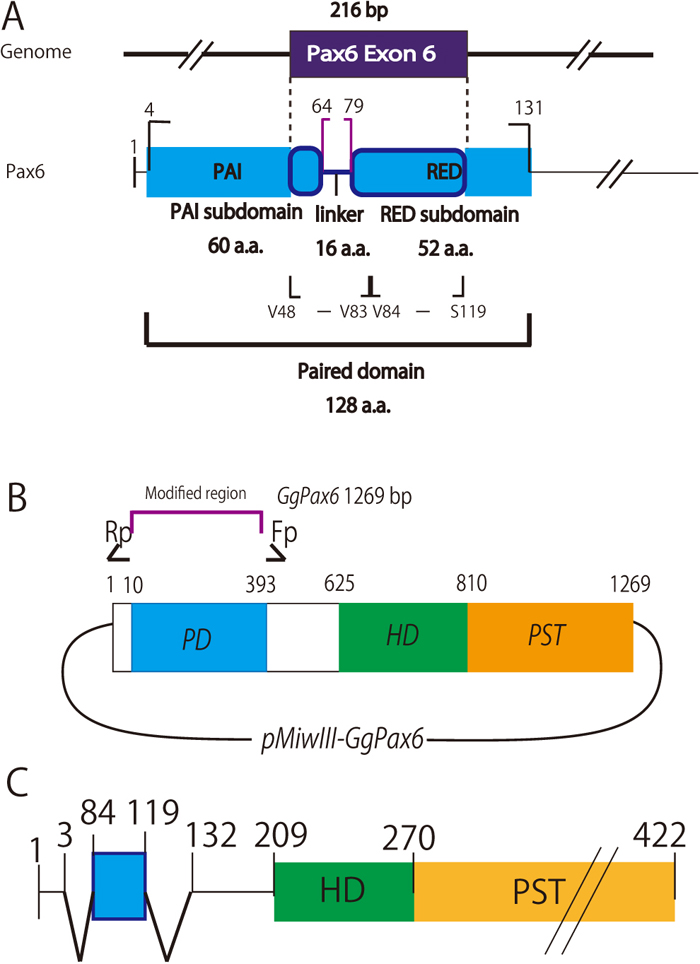
Schematic representation of the chick Pax6 paired domain and preparation of an expression plasmid encoding PD-deficient chick Pax6 but including potential NLSs of the PD. A. Exon 6 of chick Pax6, which corresponds to exon 5 of quail Pax6, encodes part of the PAI subdomain followed by the linker amino acid sequence and part of the RED subdomain (Xu et al., 1999). B. Schematic drawing of a plasmid encoding PD-deficient Pax6. Inverse PCR was carried out using primers that were designed in opposite orientations, shown as Rp and Fp (Table 1). C. A representative modified Pax6 protein (Pax6-NLSV84-S119) expressed from plasmids electroporated into the midbrain of chick embryos, where endogenous Pax6 expression is hardly detected (cf. Fig. 2A and 2B). Within the PST (proline-serine-threonine transactivation domain) region, residues N399 to the C-terminus were replaced with virion protein 16 (Hirai et al., 2010), which has robust transactivation activity (not shown because this sequence does not affect protein localization in cells).
As noted above, exon 5 of Pax-QNR (Carrière et al., 1993, 1995) corresponds to exon 6 of chick (Gallus gallus domesticus) Pax6 (gene ID: XM_015286139; protein ID: XP_015141625). Full-length chick Pax6 cDNA was prepared using RT-PCR with mRNA obtained from E9 chick eyes and midbrains according to Mochii et al. (1998) and was inserted into the expression vector pMiwIII, a derivative of pMiwZ (Suemori et al., 1990; Wakamatsu et al., 1997; Matsunaga et al., 2000). This expression vector includes the nucleotide sequence that encodes virion protein 16 (Hirai et al., 2010), which has a robust transactivation activity that can be utilized in other experiments but does not affect the localization of the fused protein in transfected cells. From the resulting Pax6-Vp16 construct, plasmids expressing modified Pax6-VP16 transcription factors that are deficient in the PD sequence were prepared by inverse PCR (Fig. 1B). Next, exon 6 of chick Pax6 was inserted into the same position that the PD sequence had originally occupied. Using this resultant construct, new plasmids expressing modified Pax6-VP16 transcription factors that are deficient in the PD sequence except for the N-terminal half (V48 - V83) or the C-terminal half (V84 - S119) amino acid sequences encoded by exon 6, each as candidate NLS sequences from the PD, were prepared, also by inverse PCR, as shown in Fig. 1C. These modified Pax6s were termed (ΔPD+NLSV48-V83)-Pax6-VP16 (shortened to Pax6-NLSV48-V83) and (ΔPD+NLSV84-S119)-Pax6-VP16 (Pax6-NLSV84-S119), respectively. The primers used are summarized in Table 1.
| Modified Pax6 | Template | Primer set |
|---|---|---|
| Pax6-ΔPD | pMiwIII-Pax6 | Fp - ACTGTTCTGCATGGTGGC |
| Rp - AGCGAAAAGCAACAGATGG | ||
| Exon 6 N-terminal Pax6-NLSV48-V83 | pMiwIII-Pax6 (fragment inserted into pMiwIII-Pax6-ΔPD) | Fp - TCGAAGCTTGCCACCATGCAGAACAGTGTGTCGAATGGATGTGTGA GTAAAATTTTG |
| Rp - CTAGCTAGCAACTTCGGGAGTCGCTACTCTCG | ||
| Exon 6 C-terminal Pax6-NLSV84-S119 | pMiwIII-Pax6 (fragment inserted into pMiwIII-Pax6-ΔPD) | Fp - TCGAAGCTTGCCACCATGCAGAACAGTGTAAGCAAAATAGCGCAGT ATAAACGAGAGTG |
| Rp - CTAGCTAGCACTGGGTATGTTATCGTTGGTACAGACC | ||
| Pax6-NLSV48-S65 | pMiwIII-Pax6-NLSV48-V83 | Fp - AGCGAAAAGCAACAGATGGGTGCCGAC |
| Rp - AGTAAAATTTTGGGCAGGTATTACGAAACTGGCTCC | ||
| Pax6-NLSI66-V83 | pMiwIII-Pax6-NLSV48-V83 | Fp - CCGCCACCATGCAGAACAGT |
| Rp - ATCAGGCCCAGGGCGAT | ||
| Pax6-NLSV48-V53 | pMiwIII-Pax6-NLSV48-S65 | Fp - AGCGAAAAGCAACAGATGGGTGC |
| Rp - CACACATCCATTCGACACACTGTTCT | ||
| Pax6-NLSV48-I56 | pMiwIII-Pax6-NLSV48-S65 | Fp - AGCGAAAAGCAACAGATGGG |
| Rp - AATTTTACTCACACATCCATTCGA | ||
| Pax6-NLSL57-S65 | pMiwIII-Pax6-NLSV48-S65 | Fp - TTGGGCAGGTATTACGAAACTGGC |
| Rp - ACTGTTCTGCATGGTGGCGG | ||
| Pax6-NLSI66-S74 | pMiwIII-Pax6-NLSI66-V83 | Fp - GCTAGCGAAAAGCAACAGATGGGT |
| Rp - ACTACCTCCGATCGCCCTGG | ||
| Pax6-NLSK75-V83 | pMiwIII-Pax6-NLSI66-V83 | Fp - AAGCCGAGAGTAGCGACTCCC |
| Rp - ACTGTTCTGCATGGTGGCGG | ||
| Pax6-NLSV78-V83 | pMiwIII-Pax6-NLSI66-V83 | Fp - GTAGCGACTCCCGAAGTTGCTAGC |
| Rp - ACTGTTCTGCATGGTGGCGG | ||
| Pax6-NLSC52-Y61 | pMiwIII-Pax6-NLSV48-S65 | Fp1 - TGTGTGAGTAAAATTTTGGGCAGGT |
| Rp1 - ACTGTTCTGCATGGTGGCGG | ||
| Fp2 - AGCGAAAAGCAACAGATGGGTG | ||
| Rp2 - GTAATACCTGCCCAAAATTTTACTCA | ||
| Pax6-NLSY61-A70 | pMiwIII-Pax6-NLSV48-V83 | Fp1 - TACGAAACTGGCTCCATCAGGC |
| Rp1 - ACTGTTCTGCATGGTGGCGG | ||
| Fp2 - GCTAGCGAAAAGCAACAGATGGGT | ||
| Rp2 - CGCCCTGGGCCTGATGGA | ||
| Pax6-NLSA70-A79 | pMiwIII-Pax6-NLSI66-V83 | Fp1 - GCGATCGGAGGTAGTAAGCCGAG |
| Rp1 - ACTGTTCTGCATGGTGGCGG | ||
| Fp2 - GCTAGCGAAAAGCAACAGATGGGT | ||
| Rp2 - CGCTACTCTCGGCTTACTACCTCCG |
These constructs were electroporated into developing chick midbrains at HH 9-12 stages (Hamburger and Hamilton, 1992), at which the expression of endogenous Pax6 can hardly be detected (Fig. 2A and 2B). Localization of the modified Pax6 in nuclei was determined by observing co-localization of the signals of DAPI (to identify nuclei) and Pax6, which was detected by an antibody against that protein. Because we first found that the N-terminal half of the putative NLS (V48 - V83) had a higher activity to drive Pax6-NLSV48-V83 into nuclei, to obtain a more detailed map of the NLS of the PD, the N-terminal half (V48 - V83) encoded by exon 6 was scanned with 6-9-amino acid windows since NLSs are usually only seven to 10 residues in length, as mentioned above. Each expression vector was prepared by inverse PCR (Table 1). Representative examples are shown in Fig. 2. We found that the chick PD-NLS activity of Pax6 broadly resides over a 36-amino acid sequence between V48 and V83 (Fig. 3) compared to a lower NLS activity shown by the C-terminal half of the putative NLS (V84-S119). The amino acid sequence between V48 and V83 mainly corresponds to the linker region of the two subdomains in the PD, namely PAI and RED, and to the C- and N-terminal amino acid residues of the PAI and RED subdomains, respectively (Fig. 4). This linker region has been reported to have no specific function between these subdomains (Xu et al., 1999). According to the crystal structure of the human Pax6 PD-DNA complex reported by Xu et al. (1999), this region corresponds to the third α-helix in the PAI subdomain, followed by the linker and four amino acid residues of the fourth α-helix in the RED subdomain (Fig. 4). The central part of the linker region contacts the minor DNA groove according to crystallographic analysis (Xu et al., 1999). Within this relatively broader NLS activity region, L57 to S65 and I66 to S74 showed higher NLS activities (Fig. 4). This consecutive region corresponds to the C-terminal half of the third α-helix (L57 to T63) and 11 amino acid residues (G64 to S74) of the linker, which consists of a total of 16 amino acid residues (G64 to A79). We have not hitherto been able to detect a typical NLS (Kosugi et al., 2009) in this region, although positively charged amino acids needed to form a NLS are distributed within it.
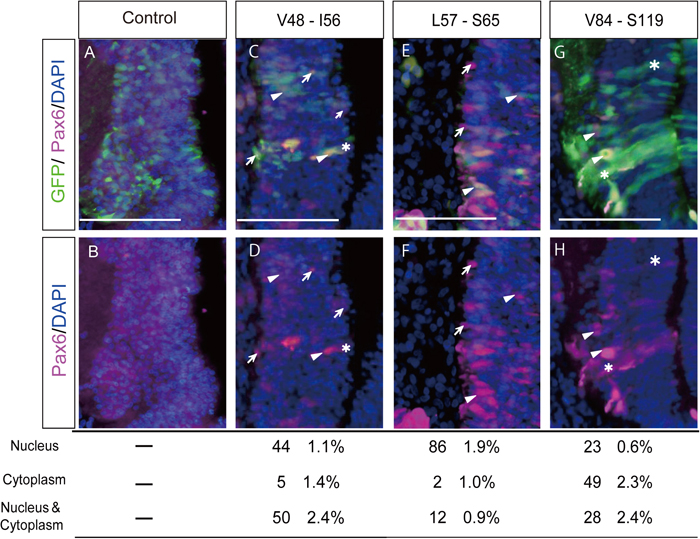
Transfection into the midbrain of modified chick Pax6 constructs harboring fragments of exon 6 encoding the putative NLS. Modified Pax6 constructs in pMiwIII were co-electroporated with pCAGGS-EGFP into the midbrain (neural tube) of chick embryos at stages 9 to 12. In ovo electroporation was carried out as described previously (Nishihara et al., 2012), modified according to Tsukiji et al. (2009). In brief, white Leghorn chicken eggs were purchased from Yamagishi (Ogaki, Japan) and incubated at 38 ℃ until they reached stages 9 – 12 (HH9-12, corresponding to the OV (optic vesicle) stages), according to Hamburger and Hamilton (1992). The plasmid solution was then injected into the neural tube (left side). An anode (0.5 mm in diameter, 1.0 mm in length; Unique Medical Imada, Tokyo, Japan) and a cathode (tungsten needle) were placed on the outside and inside of the embryo, respectively, across the OV (and surface ectoderm). Rectangular pulses (7 V, 30 ms) were then charged twice using an electroporator (BEX, Tokyo, Japan). The concentrations of plasmids were 2 μg/μl and 0.5 μg/μl for pMiwIII-modified Pax6 and pCAGGS-EGFP, respectively. After 60 h incubation, embryos were fixed with 10% formaldehyde in PBT (0.1% Tween20 in PBS) for 4 h at 4 ℃. After washing in PBT, the tissues were equilibrated in 30% sucrose in PBS, embedded in OCT compound and sectioned using a cryostat (Leica CM3050) at 7 μm. Sections were washed in TBST (0.1% Tween 20 in TBS), incubated for 30 min in 2 N HCl and washed again in TBST, and then blocked with 0.5% skim milk in TBST for 30 min. Immunostaining was performed according to the method of Tsukiji et al. (2009). A monoclonal antibody against Pax6 (1:500, Developmental Studies Hybridoma Bank, University of Iowa, USA) was used as the primary antibody. Each specimen was incubated with the primary antibody diluted with 3% BSA in PBS for 2 h at room temperature or overnight at 4 ℃. Alexa Fluor 594 (1:500, Thermo Fisher Scientific, Waltham, USA) goat anti-rabbit IgG (H + L) was used as the secondary antibody. For each modified Pax6 cDNA, more than 60 electroporated cells, identified by the expression of co-electroporated pCAGGS-EGFP, were observed and categorized according to the subcellular localization of Pax6 (mostly expressed from the electroporated construct), which was visualized with the anti-Pax6 antibody. The categorized cells are expressed as a percentage of the total cells observed. Representative results are shown with statistical data. A and B. As a negative control, a plasmid harboring a frame-shifted chick Pax6 cDNA, which results in no exogenous Pax6 expression, was electroporated into the midbrain. Endogenous Pax6 expression was hardly detected. C and D. Modified Pax6 encoding the C-terminal part of the PAI subdomain (residues V48-I56) that corresponds to the N-terminal amino acids encoded by exon 6 of chick Pax6. The proportion of transfected cells in which signals of the construct were localized in the nucleus was 44 ± 1.1% (mean ± S.D.), and that in the cytoplasm was 5 ± 1.4% (N > 3.). E and F. The highest score for the proportion of nuclear-localized signals was obtained when amino acid residues from L57 to S65 were included in the modified Pax6. Nuclear localization was detected in 86 ± 1.9% of the electroporated cells, and cytoplasmic localization was detected in 2 ± 1.0% of the electroporated cells. G and H. As a reference, a modified Pax6 construct that encodes 36 amino acid residues (residues V84-S119) was electroporated into the midbrain. Nuclear localization of the modified Pax6 was seen in 23 ± 0.6% of the electroporated cells. Exclusively cytoplasmic localization of this Pax6 was detected in 49 ± 2.3% of the cells electroporated. Arrows: cells in which Pax6 expression was detected only in nuclei; arrowheads: cells in which signals were detected in both the nucleus and the cytoplasm; asterisks: cells in which signals were detected only in the cytoplasm. Scale bars = 100 μm.
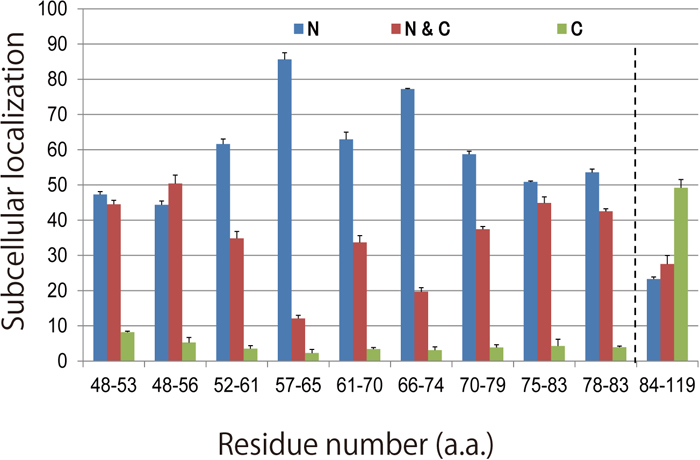
Subcellular localization of modified Pax6 proteins. Each modified chick Pax6 construct that was electroporated has its own DNA fragment that encodes a putative NLS, indicated as shown in the legend for Fig. 2. Transfected cells are categorized according to subcellular localization as described in the legend for Fig. 2. N > 3. N: nucleus; N & C: nucleus and cytoplasm; C: cytoplasm.
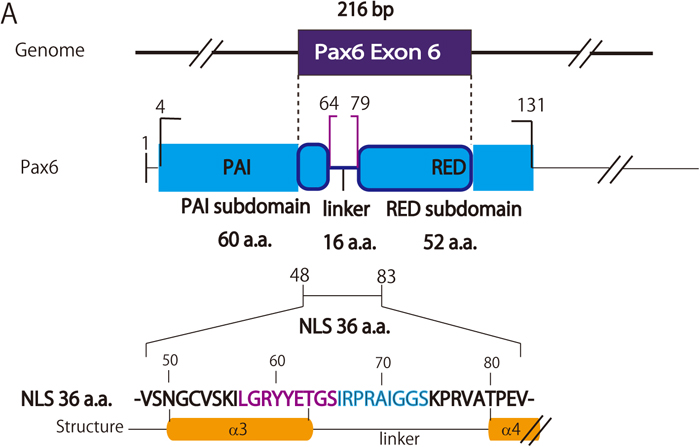
Schematic representation of the PD-NLS. PD-NLS activity broadly resides between V48 and V83, covering the N-terminal half of the amino acid residues encoded by exon 6. In this region, the higher NLS activity segments were found consecutively between L57 and S74 (colored magenta and blue, corresponding to the fragments 57-65 and 66-74, respectively, shown in Fig. 3), which spans half of the third α-helix and about 3/4 of the linker.
In the well-known NLS for the HD, the N-terminal amino acid segment LKRKLQR (HNLS5) adjacent to the HD and the Karyopherin 13 (importin 13)-binding site of the HD are required for the import of Pax6 and other paired-type homeodomain proteins into nuclei (Ploski et al., 2004). In contrast to the short HNLS5 with lower NLS activity, our results suggest that the two consecutive amino acid sequences (L57 to S65 and I66 to S74) with higher NLS activities and the surrounding amino acid residues with moderate NLS activities, together comprising the PD-NLS of Pax6, result in a higher activity of the NLS in the PD than that of HNLS5. We note that there may be two distinguishable ‘elements’ responsible for the higher PD-NLS activity between L57 and S74, because Y61 to A70 showed lower NLS activity (Fig. 3). In any case, because the range of highly active PD-NLS has been narrowed down in our study, it is now possible to further elucidate the molecular mechanism(s) by which the PD-NLS contributes to the nuclear localization of Pax6, by, for example, identifying where Karyopherin 13 binds and whether a nuclear import adaptor protein is required.
In summary, the NLS in the PD of chick Pax6 has been clarified in ovo to have a structure consisting of a relatively long amino acid sequence (36 residues, V48 through V83). This region spans the C- and N-terminal amino acid sequence of the PAI and RED subdomains in the PD, respectively, and the linker region between these subdomains (shown schematically in Fig. 4). Within this long NLS motif, the central area of 18 amino acids consisting of two consecutive nine-amino acid segments showed higher NLS activity; this area corresponds to the C-terminal half of the third α–helix of the PAI subdomain and the subsequent 11/16 amino acid residues of the linker between these subdomains (between the third and fourth α–helices). Therefore, when Pax6 is transported from the cytoplasm to the nucleus, it is possible that the PD-NLS works as a stronger NLS than the HD-NLS. Moreover, according to crystallographic analysis (Xu et al., 1999), it plays an important role to form the third α-helix of the PAI subdomain that binds to the major groove of the DNA double helix, and also consists of most of the linker between the PAI and RED subdomains that lie in the minor DNA groove and extensively contact the DNA.
Finally, concerning the 18 amino acid residues with high NLS activity, it should be noted that seven missense mutations of human PAX6 (T63P, G64V, I66N, P68S, G72S, G73D and S74G) associated with congenital eye diseases, such as aniridia, cataract, glaucoma, nystagmus, strabismus, morning glory disc anomaly and foveal hypoplasia, have been mapped within the corresponding human PD-NLS region (L57 to S65 and I66 to S74) of Pax6 (http://lsdb.hgu.mrc.ac.uk/variants.php?action=search_unique&select_db=PAX6). These amino acid residues are conserved in the PD-NLS of chick Pax6 and most of them reside in the linker region between the PAI and RED subdomains. Among these missense mutations, Azuma et al. (2003) reported that the P68S mutant human PAX6 protein (PAX6-P68S) failed to repress the promoter of PAX2, one of the target genes of PAX6. In addition, Chao et al. (2003) reported that the PAX6-G73D missense mutant protein possessed significantly reduced transcriptional activation ability. It will be intriguing to observe the efficiency of nuclear localization of these PAX6s. Interestingly, these seven amino acid residues are conserved in Pax1 and Pax9, both of which are devoid of the HD (Paixão-Côrtes et al., 2015). Therefore, it is possible that these seven residues also play, at least partly, an important functional role in these HD-deficient Pax proteins. These reports and our present results suggest the functional importance of the PD-NLS for Pax proteins to exert their biological functions.
Our results help us to further understand the molecular mechanism involved in ontogeny, where Pax6 plays a pivotal role.
We thank Drs. Toshitaka Ikeuchi, Yukio Mukai and Fang-Sik Che at the Nagahama Institute of Bio-Science and Technology for helpful discussions. Our study is indebted to Drs. Shintaro Nomura, Nobuo Nagai and the late Akitsugu Yamamoto at the same Institute for their technical advice. We thank Dr. Ian Smith for his critical reading of our manuscript. This work was partly supported by a grant from the Japan Society for the Promotion of Science to D. N. (10J07029, http://www.jsps.go.jp/), and a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan, to H. Y. (17370001).