2018 Volume 93 Issue 3 Pages 111-118
2018 Volume 93 Issue 3 Pages 111-118
In this study, we investigated the chromosome breakage caused by gametocidal (Gc) chromosome 3Ct and its interaction with the suppressor gene Igc1 (inhibitor of gametocidal gene 1) on wheat chromosome 3B. We demonstrated cytologically that patterns of 3Ct-induced chromosomal fragmentation in microspores differed from patterns observed for other Gc genes. Uninuclear microspores of the monosomic 3Ct addition line had high frequencies of micronuclei, possibly explaining its low fertility. Chromosome fragmentation was observed in prometaphase and metaphase of the first pollen mitosis in the monosomic 3Ct addition line. Patterns of chromosome fragmentation were different from those previously reported for Gc chromosomes 2S of Aegilops speltoides, 4Ssh of Ae. sharonensis and 2Ccy of Ae. cylindrica; many chromosome fragments were observed in prometaphase of the first pollen mitosis in the monosomic 3Ct addition plants. In anthers at the binuclear stage, many microspores at the uninuclear stage coexisted with normally developed microspores.
Chromosomes with gametocidal (Gc) factors induce chromosome breakage in gametes lacking them. Derived from the wheat relatives Aegilops spp., Gc genes have been identified on the chromosomes of homoeologous groups 2, 3 and 4. To date, seven genomes, namely S-related S, Sl and Ssh genomes, C-related Cc, Ccy and Ct genomes, and M-related Mo genome, of the genus Aegilops have been found to carry Gc genes (Endo and Tsunewaki, 1975; Maan, 1975; Endo, 1978; Endo and Katayama, 1978; Feldman and Strauss, 1983; Friebe et al., 1999). Chromosome 3Ct from Aegilops triuncialis L. (2n = 4x = 28, genome constitution UtUtCtCt) was the first reported Gc chromosome in wheat (Triticum aestivum L., 2n = 6x = 42, AABBDD) that can induce gamete abortion (Endo and Tsunewaki, 1975). Alloplasmic wheat lines with Ae. triuncialis cytoplasm exhibited reduced pollen and seed fertility when an acrocentric alien chromosome, Gc chromosome 3Ct, was monosomically present in the sporophyte (Endo and Tsunewaki, 1975). Homoeology of the Ae. triuncialis-derived Gc chromosome was unknown until the discovery of a normally growing disomic substitution line, DS(3B)3Ct (Endo and Katayama, 1976). Subsequently, the Gc gene on 3Ct was designated as Gc3-Ct1 (Tsujimoto, 1995; Endo, 2007).
A gene in the wheat genome that modifies the action of Gc factors has been identified. In the genetic background of ‘Norin 26’ (N26) wheat, both male and female gametes of monosomic 3Ct addition plants were fully fertile (Endo, 1978). An inhibitor gene (Igc1) against 3Ct Gc action was later found that recovered female gamete fertility almost entirely (Tsujimoto and Tsunewaki, 1985a). Tsujimoto and Tsunewaki (1985a) mapped the Igc1 locus to chromosome 3B of N26 by monosomic analysis. In progeny of plants heterozygous for Igc1 and hemizygous for 3Ct, Tsujimoto and Tsunewaki (1985b) cytologically identified aberrant chromosomes, such as telosomes and dicentrics. Recent work has mapped Igc1 to the pericentromeric region of chromosome 3B, suggesting that the gene is a homoeologous counterpart of Gc3-Ct1 that has lost Gc function (Yamano et al., 2010), as Gc3-Ct1 is also located in the pericentromeric region of 3Ct (Endo, 2007). This conclusion had been earlier inferred based on chromosome homoeology (Tsujimoto and Tsunewaki, 1985a). Gametocidal-factor modifiers in the wheat genome have also been found for chromosome 2Cc of Ae. cylindrica Host. (2n = 4x = 28, CcCcDcDc) (Endo, 1979). Although 2Cc induces no gamete abortion but chromosome structural changes in the ‘Chinese Spring’ (CS) wheat background, it induces severe abortion in ‘Jones Fife’ wheat (Endo, 1988a). The genetic factor(s) interacting with Gc chromosome 2Cc are yet to be identified. Early work also inferred the possible presence of a Gc-inhibiting modifier of the Gc2-4Ssh gene derived from Ae. sharonensis Eig (2n = 2x = 14, SshSsh) on chromosome 4B (Endo, 1988b). This modifying action was later directly attributed to a loss-of-function mutant allele of Gc2-4Ssh that inhibited chromosome breakage in heterozygous plants (Friebe et al., 2003). These data suggest that interactions between functional and nonfunctional Gc genes suppress chromosome breakage. We currently do not know the mechanisms underlying Igc1 inhibition of 3Ct Gc action.
In this study, we aimed to describe the pattern of chromosome breakage induced by the Gc chromosome 3Ct based on cytological observation of male gametogenesis. Using chromosome-specific molecular markers, we examined the transmission of chromosome 3B of both CS wheat (hereafter 3BCS) and N26 wheat (hereafter 3BN26), and of chromosome 3Ct, to evaluate the influence of genetic background (e.g., Igc1 genotype) on transmission frequency of monosome 3B. We examined the correlation between chromosome structural aberrations and presence/absence of chromosome 3Ct or 3BN26 in the offspring of 3Ct-hemizygous plants.
To examine Gc action of 3Ct and Igc1 inhibitory effects (Tsujimoto and Tsunewaki, 1985a; Yamano et al., 2010), we used the following six wheat lines: CS (2n = 42), euploid T. aestivum ‘Chinese Spring’; DA3Ct (2n = 44), disomic 3Ct addition of CS; MA3Ct (2n = 43), monosomic 3Ct addition of CS; DS(3BCS)3Ct (2n = 42), disomic substitution (3B)3Ct of CS where chromosome 3B was substituted with 3Ct; MS(3BCS)3Ct (2n = 42), monosomic substitution (3B)3Ct of CS; and F1 DS(3BCS)3Ct × N26 (2n = 42), F1 of DS(3BCS)3Ct × N26. We then scored the proportion of normal pollen grains in anthers at the uninuclear stage, the proportion of mature pollen grains in their anthers at the dehiscence stage and the selfed-seed fertility. F1 DS(3BCS)3Ct × N26, MA3Ct and MS(3BCS)3Ct were self-pollinated for analysis of transmission of 3B and 3Ct to progeny. Plants were grown in glasshouses located at the Graduate School of Agriculture, Kyoto University (N35.031, E135.79). All plant materials were provided by the National BioResource Project Wheat, Japan (http://www.shigen.nig.ac.jp/wheat/komugi/top/top.jsp).
Selfed-seed fertilityWe examined selfed-seed fertility during 2016–2017 for five to nine spikes per line. Selfed-seed fertility was scored as previously described (Yamano et al., 2010) with minor modifications. Briefly, the number of seeds in the first and second florets was counted for ten spikelets excluding the two spikelets at the top and bottom of a given spike. The fertility ratio was calculated as the number of seed-bearing florets divided by the number of counted florets. Data were analyzed with Tukey’s HSD test (α = 0.05) in R studio (https://www.rstudio.com/).
Cytological observation of male gametogenesisOne anther per floret was used to determine gametogenesis stages. The remaining two anthers were fixed in a 1:3 acetic acid to ethanol (99.5%) solution for 3 days at room temperature, and then stored at 4 ℃. Anthers were stained with acetocarmine and squashed under a cover slip. Pollen grains were observed using a BX41 optical microscope (Olympus, Tokyo, Japan). Pollen grains with two sickle-shaped sperm nuclei were scored as mature; others were scored as immature (Supplementary Fig. S1). At least 500 pollen grains of three to five anthers from different spikes per line were examined for microspores at the uninuclear and dehiscence stages. Data were analyzed with Tukey’s HSD test (α = 0.05) in R studio.
DNA extraction and PCRTotal genomic DNA was extracted from leaves of MA3Ct, F1 DS(3BCS)3Ct × N26 and MS(3BCS)3Ct progeny, following published procedures (Yabe et al., 2014) with minor modifications. We used the PLUG (PCR-based landmark unique gene) marker TNAC1648, as well as the ISBP (insertion site-based polymorphic) markers cfp1125 and cfp3310, for 3Ct and 3B. The PLUG marker is assigned to group-3 short arm’s distal-most bins (3AS4-0.45-1.00, 3BS8-0.78-1.00, 3DS4-0.59-1.00) in hexaploid wheat (Ishikawa et al., 2007), while the ISBP markers are located in bins C-3BS1-0.33 and C-3BL2-0.22, respectively (Paux et al., 2008).
Chromosome preparation and FISH (fluorescence in situ hybridization)We followed existing chromosome preparation and FISH procedures described by Sakai et al. (2009). Acetocarmine-stained chromosome images were captured with a CCD camera (DP27, Olympus). Chromosomes were stored at −80 ℃ until FISH analysis. Oligo-pSc119.2-1 (McIntyre et al., 1990) and genomic DNA of Ae. caudata L. (2n = 2x = 14, CC) were selected as probes. Genomic DNA (1 μg) was labeled with digoxigenin using DIG High Prime (Roche, Basel, Switzerland) following the manufacturer’s protocol. Oligo-pSc119.2-1 (Tamra-5′-CCGTT TTGTG GACTA TTACT CACCG CTTTG GGGTC CCATA GCTAT-3′) was synthesized and 5′-end-labeled with TAMRA (Thermo Fisher Scientific, Waltham, MA, USA). Approximately 30–50 ng of probes and 500 ng of competitor CS genomic DNA were hybridized simultaneously per slide. To detect DIG-labeled probes, we used FITC-conjugated anti-digoxigenin mouse IgG (Roche). A CCD camera (DP30BW, Olympus) captured FISH images as individual pictures of each color (blue, DAPI; green, FITC; red, TAMRA) and they were merged using ImageJ 1.51n in Fiji (Schindelin et al., 2012).
As expected for meiosis with monosomes, pollen mother cells (PMCs) of MA3Ct at metaphase I typically exhibited a 3Ct univalent (97 of 109 PMCs observed) (Fig. 1A), which was often (60 of 109 PMCs observed) associated with a univalent of an unidentified non-3Ct chromosome. At anaphase I, we observed lagging chromosomes and precocious separation of homologs (Fig. 1B). At telophases I and II, we observed micronuclei that presumably originated from the lagging chromosomes (Fig. 1C and 1D). Most uninuclear microspores (94.3%, 837 of 888 microspores observed) from an anther were well stained, and 27.5% (244 of 888) of cells possessed a micronucleus (Fig. 1E). Chromosome breakage patterns in the first pollen mitosis of MA3Ct differed from those reported for Gc1 or Gc2 genes in which we observed many chromosome fragments of various sizes in the middle of the spindle in anaphase of the first pollen mitosis. These fragments consisted of similar-sized chromatids, indicating that breakage occurred before S-phase of the first pollen mitosis (Nasuda et al., 1998). In this study, however, we observed condensed chromosome fragments at prometaphase and metaphase in MA3Ct (Fig. 1F and 1G), which were not found in the Gc1 and Gc2 systems. In more developmentally advanced anthers, numerous microspores at the uninuclear stage were present together with normally developed binuclear microspores (Fig. 1H and 1I).

Male gametogenesis in the monosomic 3Ct addition line of ‘Chinese Spring’ wheat. (A) Meiotic metaphase I with one univalent. Arrow indicates a 3Ct univalent. (B) Meiotic anaphase I with precociously separated chromatids. (C) Meiotic telophase I with one micronucleus in each daughter cell. (D) Tetrad consisting of two cells (lower) with a micronucleus and two cells (upper) without. (E) Uninuclear microspores; one possessing a micronucleus is indicated with an arrow. (F and G) Chromosome fragmentation observed in a microspore in a stage prior to anaphase of first pollen mitosis. (H) Anaphase (arrow) and prophase (arrowhead) microspores of first-pollen mitosis. (I) Binuclear microspores; the arrow indicates a normal microspore, and the arrowhead indicates an arrested microspore. Scale bars = 20 μm.
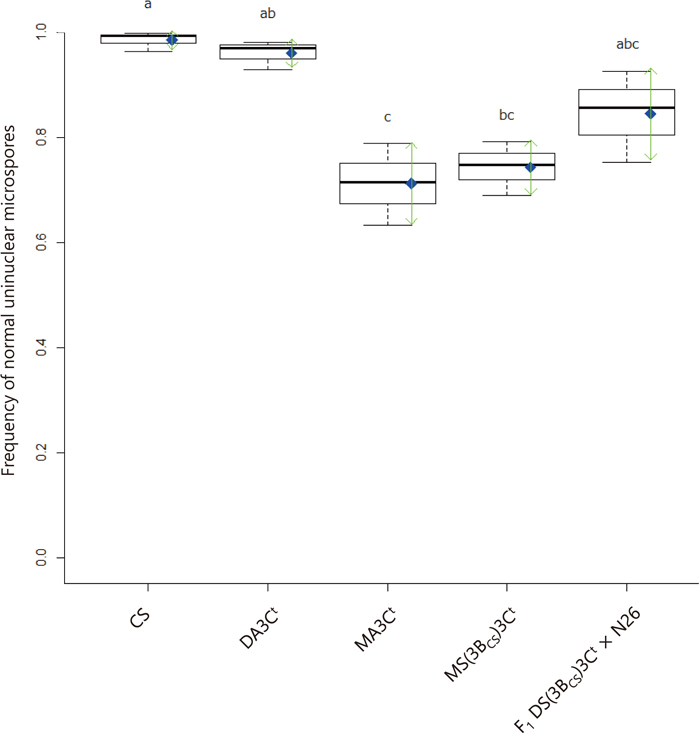
Boxplots indicating frequencies of normal uninuclear microspores across five lines that differ in 3Ct doses. Thick horizontal bars indicate medians. Blue diamonds indicate means, and green arrows indicate standard deviations. Different letters represent significant differences at P < 0.05 via Tukey’s HSD test.
Gc monosomic plants can suffer from severe sterility due to the loss of the Gc monosome during meiosis (Tsunewaki et al., 1996). As expected, the average selfed-seed fertility of MA3Ct was 13.0% in this study, which was as low as the fertility (15%) previously reported by Tsujimoto and Tsunewaki (1985a). We presented the cytological evidence of chromosome loss that 27.5% (244 of 888) of uninuclear microspores in MA3Ct possessed micronuclei.
Selfed-seed fertility and proportion of mature pollen in 3Ct monosomic plantsThe average selfed-seed fertility was 95.6 (N = 8 spikes examined), 81.1 (N = 6) and 83.0% (N = 5) for CS, DA3Ct and DS(3BCS)3Ct (control) lines, respectively. Fertility was significantly lower (P < 0.01) in 3Ct monosomic lines (13.0% (N = 5) for MA3Ct and 15.0% (N = 5) for MS(3BCS)3Ct) than in CS. Selfed-seed fertility of F1 DS(3BCS)3Ct × N26 (88.3%, N = 3) was similar to that of the 3Ct disomic controls (DA3Ct and DS(3BCS)3Ct). Indeed, the frequency of mature pollen just before dehiscence in F1 DS(3BCS)3Ct × N26 was significantly different from those in CS and DA3Ct (Fig. 3).
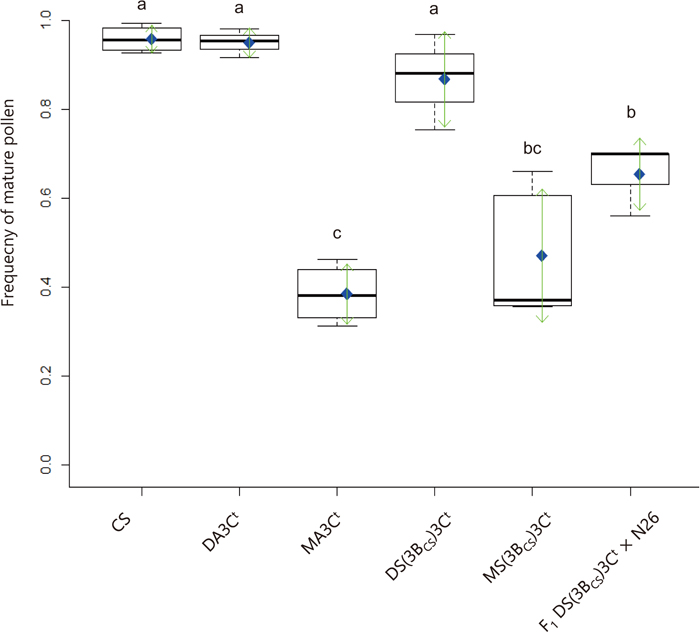
Boxplots of frequencies of mature pollen in anthers of six wheat lines at anthesis. Thick bars indicate medians. Blue diamonds indicate means, and green arrows indicate standard deviations. Different letters represent significant differences at P < 0.05 via Tukey’s HSD test.
Average frequency of mature pollen in control CS and DA3Ct reached 95.7% and 94.9%, respectively. Mature pollen frequency dropped significantly in 3Ct-hemizygous lines (MA3Ct, MS(3BCS)3Ct and F1 DS(3BCS)3Ct × N26) (Fig. 3). As expected of an Igc1 carrier line, F1 DS(3BCS)3Ct × N26 had more mature pollen than MA3Ct, but notably less than CS, DA3Ct or DS(3BCS)3Ct.
Transmission rates of chromosomes 3BN26, 3BCS and 3Ct and chromosome structural aberrations found in the progeny of F1 DS(3BCS)3Ct × N26We examined 3B and 3C transmission across 58, 22 and 20 plants obtained from self-pollinated spikes of F1 DS(3BCS)3Ct × N26, MS(3BCS)3Ct and MA3Ct, respectively (Table 1). The MS(3BCS)3Ct and MA3Ct lines did not produce plants lacking 3Ct. Within the selfed progeny of F1 DS(3BCS)3Ct × N26 we found eight and 20 plants lacking 3BN26 and 3Ct, respectively. Eight of the 22 selfed progeny of MS(3BCS)3Ct plants lacked 3BCS. The MA3Ct line did not produce plants lacking 3BCS. In the F1 DS(3BCS)3Ct × N26 progeny, we observed aberrations including acrocentric, dicentric and ring chromosomes (Table 2, Fig. 4A), regardless of the presence or absence of 3BN26 or 3Ct. One plant possessed disomic 3Ct with a deletion in chromosome arm 5BS (Fig. 4B).
| Parental lines | With 3B and 3Ct (%) | Without 3Ct (%) | Without 3B (%) | Without 3B and 3Ct (%) | Total |
|---|---|---|---|---|---|
| F1 DS(3BCS)3Ct × N26 | 30 (51.7) | 20 (34.5) | 8 (13.8) | 0 (0.0) | 58 |
| MS(3BCS)3Ct | 14 (63.6) | 0 (0.0) | 8 (36.7) | 0 (0.0) | 22 |
| MA3Ct | 20 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 20 |
| 3B as indicated by cfp1125 | |||||||
|---|---|---|---|---|---|---|---|
| Present | Absent | Total | |||||
| Y | N | Y | N | Y | N | ||
| 3Ct as indicated by TNAC1648 | Present | 8 | 5 | 4 | 1 | 12 | 6 |
| Absent | 10 | 4 | 0 | 0 | 10 | 4 | |
| Total | 18 | 9 | 4 | 1 | 22 | 10 | |
Y and N denote the presence and absence of aberrant chromosome(s), respectively.
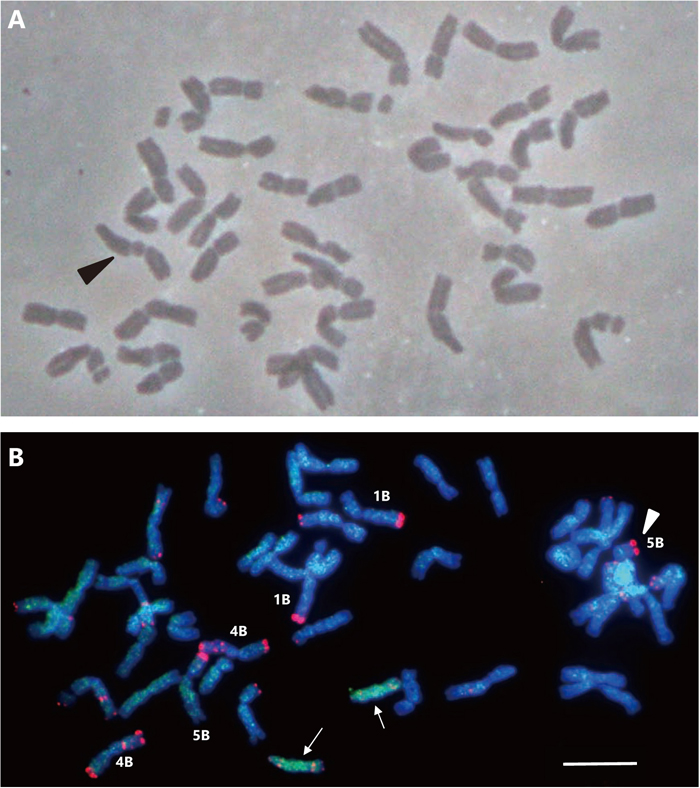
Metaphase spreads of selfed F1 DS(3BCS)3Ct × N26 progeny, indicating the occurrence of chromosome modification. (A) Metaphase spread of a cell (2n = 41) with a dicentric chromosome (arrowhead). (B) FISH image of a cell (2n = 43) with disomic 3Ct chromosomes (arrows) and a 5BS telosome (arrowhead) as represented by a strong terminal FISH signal (Fu et al., 2015). The pSc119.2 probe yields red signals and the C genome DNA probe yields green GISH signals. Chromosomes 1B, 4B and 5B with strong telomeric pSc119.2 signals are indicated by white letters. Chromosomes 1B and 5B can be distinguished by the presence of a secondary constriction on the short arm. The strong terminal pSc119.2 signals on chromosome 1B are on the long arm, while those on chromosome 5B are on the short arm. Note that 3Ct morphology is a telosome of the long arm. Scale bar = 10 μm.
The Gc chromosome 3Ct is well known to induce abnormality during gametogenesis in 3Ct-hemizygous plants, reducing male and female gamete fertility. Thus, 3Ct is preferentially transmitted to the next generation in the CS genetic background (Tsujimoto and Tsunewaki, 1985a; Endo, 1988a; Tsujimoto et al., 1990). The Gc inhibitor Igc1 was considered dominant because its presence recovers fertility of 3Ct-hemizygous plants, and gamete abortion was not observed in the N26 genetic background (Endo, 1978; Tsujimoto and Tsunewaki, 1985a). However, no study until now has investigated the molecular mechanisms of inhibitory effects of Igc1 on 3Ct Gc action.
Dominant inhibition of 3Ct Gc function by Igc1 was indicated by high selfed- and crossed-seed fertilities (90 and 89%, respectively) in the F1 progeny of the DA3Ct × N26 cross (Tsujimoto and Tsunewaki, 1985a). This result indicates that the Igc1 gene acts not only on gametes with it but also on gametes without it; the Igc1 gene sporophytically (or trans-gametophytically) inhibits 3Ct Gc function. Tsujimoto and Tsunewaki (1985a) further reported that male gametes with Igc1 have a 50% advantage over those carrying igc1 in the progeny of the cross between DA3Ct × F1 (N26 × CS). We observed that chromosome 3BN26 in F1 DS(3BCS)3Ct × N26 had a higher transmission rate than chromosome 3BCS in MS(3BCS)3Ct (Table 1). The observed difference in transmission of chromosome 3B does not contradict the previous results (Tsujimoto and Tsunewaki, 1985a). Since the present observation was made in different genetic backgrounds (F1 between CS × N26 versus CS, both with monosomes 3Ct and 3B), we cannot conclude that the Igc1 gene in combination with 3Ct ensured transmission of 3BN26. Future analysis of the transmission rates of molecular markers on chromosome 3B in test-crossed populations of DA3Ct × N26 will reveal whether chromosome 3BN26 is preferentially transmitted, or, more precisely, whether the advantage of Igc1-bearing gametes in fertilization is driven by the Igc1 gene itself or by linked genetic elements.
Gametocidal chromosomes 3Ct and 2CC have been used to systematically induce aberrations in native and alien chromosomes in wheat (see Endo, 2007 for review). In general, strong Gc action leads to gamete abortion, while incomplete Gc inhibition or mild Gc action results in chromosome aberrations only (Endo, 1990). Chromosome 3B of CS (3BCS) was reported to have an igc1 allele that partially inhibited 3Ct Gc behavior in male gametogenesis (Tsujimoto, 1986). In this study, we found a plant with disomic 3Ct and monosomic 3B, together with unidentified truncated chromosomes. If we assume that chromosomal structural aberration occurred only in plants lacking 3Ct, 3Ct disomy should have occurred through misdivision during meiosis of one parent, whereas chromosomes would have been rearranged in the other parent. Alternatively, the unusual chromosome composition might occur if we assume that chromosome aberrations are inducible in a gamete with both 3BN26 and 3Ct (Tsujimoto and Tsunewaki, 1985a; Tsujimoto, 1986). Future studies using test-crossed progeny should be performed to test whether 3Ct can structurally modify chromosomes irrespective of Igc1 presence.
We thank Dr. Miyuki Nitta for technical support and Mr. Motohiro Yoshioka for valuable comments pertaining to the research. This paper is Contribution Number 618 from the Laboratory of Plant Genetics, Graduate School of Agriculture, Kyoto University. The work was supported in part by JSPS KAKENHI (Grant-in-Aid for Scientific Research B, Grant numbers JP25292007 and JP17H03747 to S. N.).