2019 Volume 94 Issue 4 Pages 141-149
2019 Volume 94 Issue 4 Pages 141-149
The vomeronasal organ (VNO) plays a key role in sensing pheromonal cues, which elicit innate responses and induce social and sexual behaviors. The vomeronasal receptor 1 genes, V1Rs, encode members of a pheromone receptor family that are mainly expressed in the VNO. Previous studies have revealed that the V1R family shows extraordinary variety among mammalian species owing to successive gene gains and losses. Because species-specific pheromonal interaction may facilitate species-specific reproductive behaviors, understanding the evolution of V1Rs in terms of their origin, repertoire and phylogeny should provide insight into the mechanisms of animal diversification. Here I summarize recent studies about the V1R family from its initial discovery in the rat genome to extensive comparative analyses among vertebrates. I further introduce our recent findings for V1Rs in a broad range of vertebrates, which reveal unexpected diversity as well as shared features common among lineages.
The olfactory sense is essential to acquire chemical information from the external world, allowing animals to respond with appropriate behavior. Pheromones, which are secreted by an individual and perceived by another individual of the same species, are especially important for eliciting social and sexual behaviors (Karlson and Lüscher, 1959). In terrestrial vertebrates, pheromones are predominantly detected by the vomeronasal organ (VNO). The VNO is anatomically distinct from the main olfactory epithelium (MOE), which detects general odorants (Firestein, 2001; Brennan and Zufall, 2006). Neurons of the VNO (vomeronasal sensory neurons: VSNs) project to the accessory olfactory bulb (AOB), whereas those of the MOE (olfactory sensory neurons: OSNs) project to the main olfactory bulb (MOB) (Chamero et al., 2012, Fig. 1). The VSNs have a specific transduction mechanism, involving a diacylglycerol-activated cation channel and TRPC2 (transient receptor potential channel 2), which is essential for a fully functional VNO (Stowers et al., 2002).
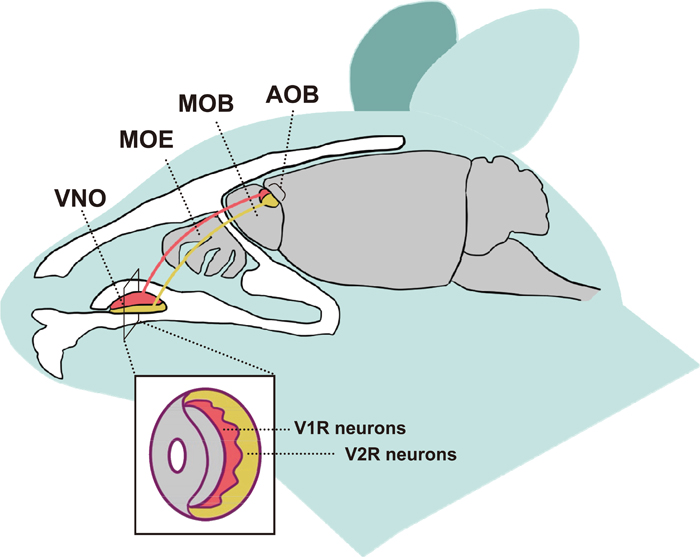
Schematic diagram of the mammalian olfactory system highlighting the VNO. Sagittal view of a mouse head showing the main olfactory epithelium (MOE), vomeronasal organ (VNO), main olfactory bulb (MOB) and accessory olfactory bulb (AOB). A coronal view of the VNO is shown in the box. In the VNO, apical VSNs express V1Rs and project their axons to the anterior (red) AOB, and basal VSNs express V2Rs and project to the posterior (yellow) AOB.
Vomeronasal receptor genes are classified into two multigene families, namely type-1 (V1Rs) and type-2 (V2Rs), which are both seven-transmembrane G protein-coupled receptors (Nei et al., 2008). These two receptor families are distinct in that the V1R-positive VSNs reside within the apical layer of the VNO, co-express Gαi2, and project to the anterior AOB, whereas V2R-positive VSNs reside within the basal layer, co-express Gαo, and project to the posterior AOB (Chamero et al., 2012, Fig. 1). Previous bioinformatic analyses for several mammalian genomes revealed that V1Rs and V2Rs show extraordinary variation in gene number and in the proportion of intact genes compared with pseudogenes (Grus et al., 2005; Yang et al., 2005; Young et al., 2005), which have been generated by species-specific gene duplication and loss (Nei et al., 2008). Consequently, size variation of the V1R repertoire exceeds that of any other multigene family (Grus et al., 2005). Given that the evolution of vomeronasal receptors may reflect the history of speciation and adaptation in a broad range of vertebrates, analyses of such genes may provide important insight into the molecular mechanisms underlying pheromone-mediated animal diversification.
In this review, I focus on the V1R family in particular, from its initial discovery to very recent findings. Recent V1R data are largely dependent on new advances in genome bioinformatics methods, which are still expanding rapidly. After the initial discovery of V1Rs in the rat genome (Dulac and Axel, 1995) they were characterized extensively in the genomes of a broad diversity of mammals (Grus et al., 2005; Young et al., 2005), frogs (Date-Ito et al., 2008) and reptiles (Brykczynska et al., 2013). Several groups revealed the extreme variability in V1R repertoires across tetrapods (land vertebrates, including mammals, amphibians, reptiles and birds) due to species-specific gene duplication and loss. V1Rs were also found in multiple species of teleost fish, which do not have VNOs, showing that they were highly conserved in these ancestral vertebrate lineages (Pfister and Rodriguez, 2005; Saraiva and Korsching, 2007). In addition to mammals and teleosts, V1Rs have been explored in the coelacanth, which was found to possess fish-type as well as tetrapod-type V1Rs (Nikaido et al., 2013). Recently, our group also found a highly divergent V1R in cichlid fishes (Nikaido et al., 2014), and a highly conserved V1R shared among most bony vertebrates (Suzuki et al., 2018). Both of these discoveries were quite unexpected and forced a reconsideration of previous ideas about V1R evolution. Further investigation of V1Rs will be essential to elucidate the general mechanism for pheromone detection systems as well as the origin of the VNO in vertebrates.
V1R genes were initially identified in the rat genome by Dulac and Axel (1995). Prior to this discovery, Buck and Axel (1991) identified a large multigene family encoding olfactory receptors specifically expressed in the MOE. They concluded that ~1,000 copies of olfactory receptor genes exist in mammalian genomes, which enables us to discriminate diverse odorant chemicals. Dulac and Axel (1995) predicted that the pheromone receptors, which may be specifically expressed in the VNO, would be distinct from the olfactory receptors. They designed an experimental strategy based on two hypotheses: 1) expression of the pheromone receptors is restricted to the VNO; and 2) individual VNO neurons express different receptor genes according to the “one neuron–one receptor” rule. Differential screening of cDNA libraries derived from individual VNO neurons identified some genes that were differentially expressed among different VNO neurons. The punctate expression patterns of the mRNA in the VNO suggested that these genes are likely to encode pheromone receptors. Based on genomic Southern hybridization assays as well as quantitative screening of genomic libraries, the authors estimated the number of putative pheromone receptor genes to be 30 or 40 copies. Since then, great advances in whole-genome sequencing technologies have allowed us to characterize these putative pheromone receptor genes for a broad diversity of mammals. This novel multigene family was named vomeronasal receptor type I (V1R) or Vmn1. The copy numbers of intact V1Rs were revealed to be 106 and 187 in rat and mouse, respectively, and are much higher than expected from their initial discovery (Nei et al., 2008).
The biological function of V1Rs has also been extensively examined. Del Punta et al. (2002) generated mutant mice in which a 600-kilobase genomic region that contains a cluster of 16 intact V1Rs was deleted. The behavioral impairment and chemosensory deficit in the mutant mice suggested a role for V1Rs as pheromone receptors. Furthermore, mutant mice deficient for Gαi2, which facilitates signal transduction by V1Rs in the apical VNO neurons, showed impairment of aggressive behaviors in males and lactation in females, implying an essential role for V1R-Gαi2 signal transduction in vomeronasal function (Norlin et al., 2003).
Since the V1R multigene family was discovered, it has been extensively characterized in a broad diversity of mammals in order to elucidate how highly diversified social and sexual behaviors have been acquired during evolution. Rodriguez et al. (2002) identified a total of 293 V1R sequences (137 intact, 60 partial, 96 pseudo) in the mouse genome, showing rapid evolution of these genes. Next, two research groups conducted a genome-wide comparison, showing the extreme variability in V1R repertoires across mammalian lineages due to species-specific gene duplication and loss (Grus et al., 2005; Young et al., 2005, 2010; reviewed in Rodriguez, 2005; Nei et al., 2008, summarized in Fig. 2). Zhang et al. (2007) compared the gene repertoires between V1Rs and olfactory receptor genes (ORs) in mice and rats, showing that V1Rs evolved more rapidly than ORs in a species-specific manner. Moreover, Kurzweil et al. (2009) and Karn et al. (2010) revealed differences in V1Rs in closely related mouse species and subspecies, respectively. These lines of evidence suggest that V1Rs contribute to the speciation and diversification of mammals.
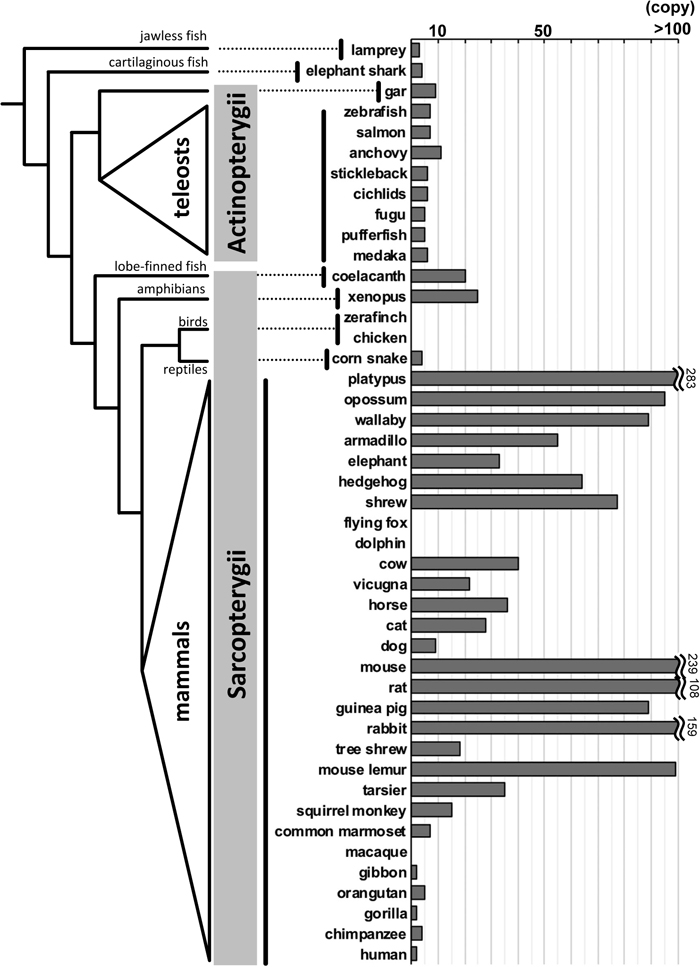
Number of intact V1Rs among a broad range of vertebrates. In the graph on the right, bars for species having more than 100 gene copies are truncated and the actual numbers are indicated. The phylogenetic tree of vertebrates with taxonomic classification names is shown on the left side. The copy number was based on the following references (Saraiva and Korsching, 2007; Date-Ito et al., 2008; Grus and Zhang, 2009; Young et al., 2010; Ota et al., 2012; Nikaido et al., 2013; Zapilko and Korsching, 2016; Moriya-Ito et al., 2018). Note that the V1R repertoire is highly variable in mammals, from 283 in platypus to 0 in dolphin and bat, but much less variable in teleost fish (6 copies in most species). The number of V1Rs increases dramatically in species belonging to the Sarcopterygii.
Grus et al. (2005) also proposed that the number of intact V1Rs correlates with the anatomical complexity of the VNO. This hypothesis is consistent with the absence of intact V1Rs in whales, dolphins and some bats (Young et al., 2010; Kishida et al., 2015), which do not possess a VNO. The decline of the number of intact V1Rs is also evident in anthropoid primates (hominoids, Old World monkeys and New World monkeys; Yoder and Larsen, 2014; Moriya-Ito et al., 2018, Fig. 2), in which the VNO is anatomically oriented to be reduced or vestigial (Smith et al., 2014). Only one apparent exception was observed in the dog genome (Boxer dog), in which the number of intact V1Rs is unexpectedly small in view of the functional VNO and a complex system of pheromone-based behaviors in wild canine species (Young et al., 2005, Fig. 2). Interestingly, several intact V1Rs have been characterized in humans and were shown to be expressed in the MOE (Rodriguez et al., 2000) even though we lack an anatomically intact VNO.
Although teleost fish do not have a VNO, they have V1Rs. Pfister and Rodriguez (2005) revealed the existence of a mammalian V1R-like sequence in multiple species of teleost fish. Subsequently, extensive bioinformatic analyses revealed that a total of six highly divergent V1Rs exist in five model fish species: zebrafish, medaka, stickleback, fugu and pufferfish (Saraiva and Korsching, 2007; Pfister et al., 2007). The teleost V1Rs were also called ORAs (olfactory receptor class A) because they are expressed in the olfactory epithelium (OE), which is not differentiated into the MOE and VNO. In addition to the above five fish species, V1Rs were also characterized in lamprey (Grus and Zhang, 2009; Libants et al., 2009), elephant shark (Grus and Zhang, 2009), cichlids (Ota et al., 2012), eel (Churcher et al., 2015), salmon (Johnstone et al., 2012) and anchovy (Zhu et al., 2016). The presence of V1Rs in lamprey and elephant shark indicates that their origin is old in aquatic vertebrate evolution (Grus and Zhang, 2009). Indeed, the phylogenetic positions of three V1Rs of lamprey, the basal lineage of all vertebrates, are close to V1R3 or V1R4 of teleost fish or a much older clade (Fig. 3), implying that vertebrate V1Rs originated in these ancestral clades.
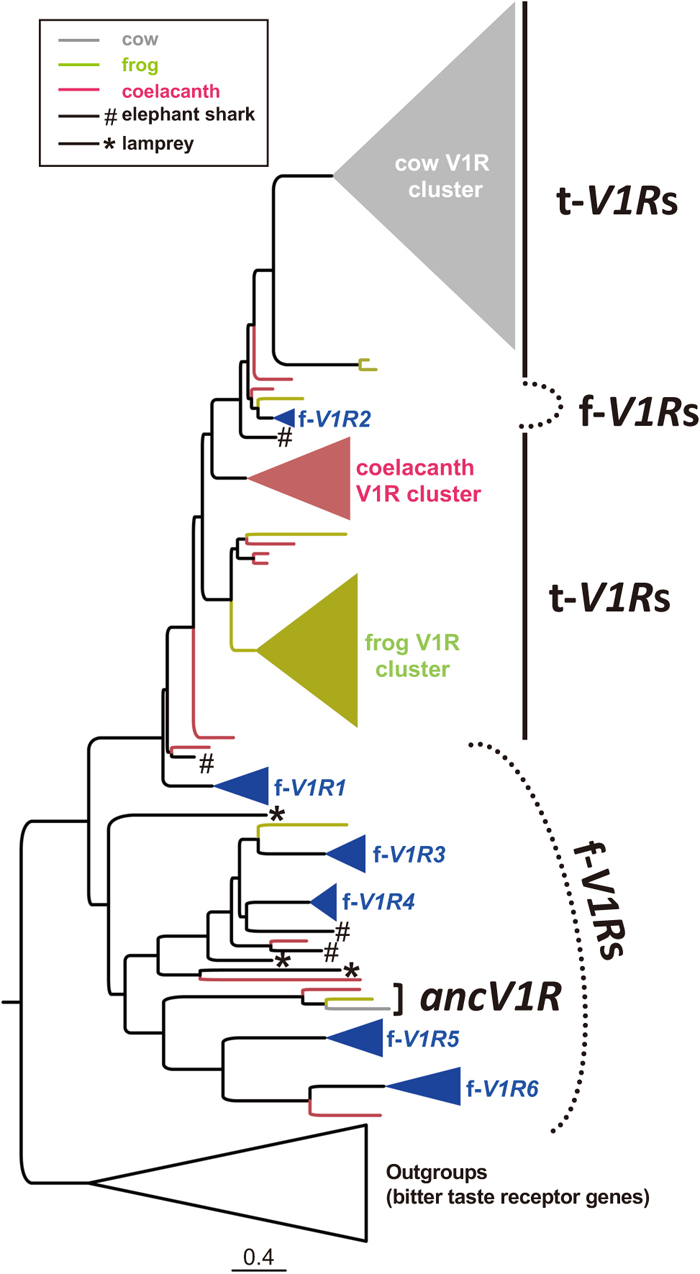
Evolutionary relationships of t-V1Rs, f-V1Rs and ancV1R. The maximum likelihood tree of the amino acid sequences of V1Rs of 10 representative vertebrates under the JTT+G+F model constructed by RAxML 8.2.4 (Guindon and Gascuel, 2003). The scale bar indicates amino acid substitutions per site. The species are indicated as follows: cow (gray), frog (green), coelacanth (red), five teleost fishes – zebrafish, stickleback, fugu, pufferfish and medaka – (blue), elephant shark (hash #), sea lamprey (star *). The bitter taste receptor type 2 sequences of vertebrates were used for outgroups of V1Rs. Note that the f-V1Rs originated earlier than t-V1Rs, which are derived from the common ancestor of f-V1R1 and f-V1R2. Importantly, an orthologous relationship is shared among a broad range of species including cow, frog and coelacanth for ancV1R but not for t-V1Rs or f-V1Rs. Recently, ancV1R was shown to be shared among most bony vertebrates (Suzuki et al., 2018).
Although the V1Rs of mammals and fish belong to the same gene family, they are distinct in terms of the diversity of their repertoires. Whereas the repertoires of V1Rs are highly diverse in mammals, they are conserved among teleost species. Indeed, most teleosts possess a set of six orthologous V1Rs except for some gene gain or loss in pufferfish, salmon and anchovy (Fig. 2). In addition to this repertoire, each of the V1Rs is highly conserved among teleost fishes at the DNA sequence level (Saraiva and Korsching, 2007; Ota et al., 2012). For example, comparisons of the V1R sequences among distantly related rockfishes (Johansson and Banks, 2010) as well as nine salmonid species (Johnson and Banks, 2011) revealed that they are all highly conserved among species. Although the ligands for fish V1Rs remain largely unexplored, the high level of conservation of teleost V1Rs in repertoires as well as in sequence implies that they recognize a very small number of ligands, which are common across a broad range of fish species. Johansson and Banks (2010) proposed that the fish V1Rs are involved in assessing mate condition, gender, reproductive status and other fitness-related traits, which are important across multiple species regardless of species specificity.
In addition to mammals and teleosts, V1Rs have been further explored in other vertebrates. Given that terrestrial vertebrates experienced a massive environmental transition from water to land, their V1Rs are also expected to have changed their ligands during that period. Indeed, Shi and Zhang (2007) have characterized and compared the V1Rs as well as other chemoreceptor genes among 11 vertebrates (mammals, chicken, frog and teleosts), proposing that the V1Rs increased in the frog (Fig. 2), in which a morphologically identifiable VNO first appeared during evolution. Saraiva and Korsching (2007) also revealed that frog V1Rs, which are phylogenetically close to mammalian V1Rs, increased in a species-specific manner. They suggested that the expansions of these V1Rs is related to the origin of the VNO in the common ancestor of tetrapods (Shi and Zhang, 2007). Hereafter, I refer to less diverse “fish-type” V1Rs as f-V1Rs and highly diverse “tetrapod-type” V1Rs as t-V1Rs. Phylogenetic analyses revealed that t-V1Rs are derived from the common ancestor of f-V1R1 and f-V1R2 (Fig. 3).
Nikaido et al. (2013) explored the V1Rs in the genome of the coelacanth, which is believed to be one of the closest extant relatives of tetrapods. Interestingly, the number of intact V1Rs (20 intact genes) of coelacanth was comparable to that of frog (Fig. 2). Indeed, the coelacanth has highly diverse t-V1Rs, suggesting that the increase of t-V1Rs occurred earlier than the emergence of tetrapods (Fig. 3, Nikaido et al., 2013). Furthermore, Zapilko and Korsching (2016) found a few t-V1Rs in the spotted gar, an early-diverging ray-finned fish, and proposed that t-V1Rs originated far earlier than expected. This earlier origin of t-V1Rs in vertebrate evolution may provide important insight into the ligands of V1Rs. Thus, the above results imply that the t-V1Rs already existed in the common ancestor of bony fishes and allowed them to detect pheromones underwater, and that subsequently these ancestral t-V1Rs were co-opted to detect pheromones in the terrestrial environment.
It is noteworthy that few or no V1Rs are found in the genomes of lizards, snakes, birds, alligators and turtles even though they are terrestrial vertebrates (Silva and Antunes, 2017). The lack of V1Rs in birds and alligators is consistent with the anatomical observations that VNOs are degenerate or vestigial in these groups (Døving and Trotier, 1998). The VNO of turtles is also equivocal (Silva and Antunes, 2017) in that it is anatomically not fully separated from the MOE (Wakabayashi and Ichikawa, 2008), which may be related to the reduction of the number of turtle V1Rs. In lizards and snakes, which possess a highly developed VNO, a transcriptome analyses of the VNOs revealed that they retain an extremely limited number of V1Rs but exhibit a large number of V2Rs (Brykczynska et al., 2013). These results imply that lizards and snakes are less dependent on V1R-mediated pheromone detection through the VNO.
De-orphanization, the process of describing an orphan receptor whose endogenous ligand has been identified, is of primary importance to elucidating the function of V1Rs. Although the ligands of most V1Rs are yet to be explored, several studies have identified possible ligands in mice and zebrafish. Boschat et al. (2002) applied electrophysiological and calcium imaging techniques to single VSNs and found that V1rb2 receptor detects 2-heptanone, which is present in mouse urine and can act as a pheromone. Isogai et al. (2011) extensively explored possible ligands for V1Rs by applying two-color in situ hybridization assays using probes against egr-1 (a marker for neuronal activation) and one specific receptor gene on VNO sections of individual mice that were exposed to variable stimuli. In this study, some V1Rs were shown to detect and discriminate distinct structural classes of sulfated steroids, which may act as sensitive reporters of the endocrine state in a broad range of vertebrates, including lamprey (Sorensen et al., 2005), goby (Murphy et al., 2001), cichlids (Cole and Stacey, 2006), tilapia (Keller-Costa et al., 2014) and amphibians (Sansone et al., 2015).
In zebrafish, Behrens et al. (2014) applied heterologous expression techniques and revealed that ORA1 (f-V1R2) recognizes 4-hydroxyphenylacetic acid, which was shown to elicit oviposition behavior. Cong et al. (2019) also applied heterologous expression techniques and showed that six ORAs (f-V1Rs) of zebrafish responded specifically to bile acids/salts, which elicit strong attraction behavior in zebrafish. Bile acids/salts are major end-metabolites of cholesterol that are released in feces and act as semiochemicals or pheromones in various fish species (Buchinger et al., 2014). At present, however, it is unclear whether the attraction behaviors elicited by the above chemicals are driven solely by V1Rs or with additional co-receptors (ORs, V2Rs). To address this question conclusively one needs to perform two-color in situ hybridization assays of the olfactory organ in zebrafish using probes against V1Rs and other marker genes for neuronal activation.
Whereas each of the f-V1R orthologs is highly conserved among distantly related fish species, our group revealed an unexpectedly large diversity of f-V1Rs in closely related cichlid species. The cichlids in the African Great Lakes have been intensely studied by evolutionary biologists because they are highly diverse in species (Kocher, 2004). Ota et al. (2012) and Nikaido et al. (2014) characterized and compared six f-V1Rs in more than 30 cichlids and found that f-V1R2 (ORA1) alleles were highly dimorphic between closely related species pairs. Evolutionary analyses revealed that these two f-V1R2 alleles emerged through a process of positive Darwinian selection, implying a functional distinctiveness of these two alleles in detecting pheromones. We thus suggested that highly dimorphic f-V1R2 led to the incipient step of population divergence and speciation. Signatures for the operation of positive Darwinian selection were further detected in f-V1R1, f-V1R3 and f-V1R6. The occurrence of such events and the unexpected gene diversity in one f-V1R family is suggestive of the contribution of f-V1Rs to species diversification in cichlids. Specification of the ligands of each f-V1R and evaluation of the relevant physiological effects may provide crucial insight into the mechanism of speciation in cichlids.
Recently, our group found a novel clade of V1R gene, named ancient V1R (ancV1R), which is shared among most bony vertebrates from the basal lineage of ray-finned fishes to mammals (Suzuki et al., 2018, Fig. 4A). Given that no ancV1R sequences were found in the genomes of elephant shark and lamprey (Fig. 3), ancV1R emerged after the divergence of cartilaginous fish (Fig. 4A). Importantly, the orthologous relationship of ancV1R has been conserved for more than 400 million years of vertebrate evolution. Conservation of orthologous relationships is quite rare in mammals, in which t-V1Rs are highly diverse and in which no genes orthologous to f-V1Rs have been found so far. Actually, although f-V1R2 shares a common ancestry with mammalian t-V1Rs, they do not have a one-to-one orthologous relationship. We also found a strong correlation between the pseudogenization of ancV1R and the loss of an intact VNO (Suzuki et al., 2018). Furthermore, ancV1R was shown to be expressed in most vomeronasal sensory neurons (VSNs) in contrast with canonical V1Rs, which show punctate patterns of expression called the “one neuron–one receptor” rule (Fig. 4B). For the first time, we showed that ancV1R is co-expressed with canonical V1Rs or V2Rs in the VNO, and proposed the new hypothesis of the “one neuron–two receptors” rule. The high degree of evolutionary conservation as well as the broad expression pattern imply an essential functional role(s) for ancV1R in VNO-mediated pheromone detection.
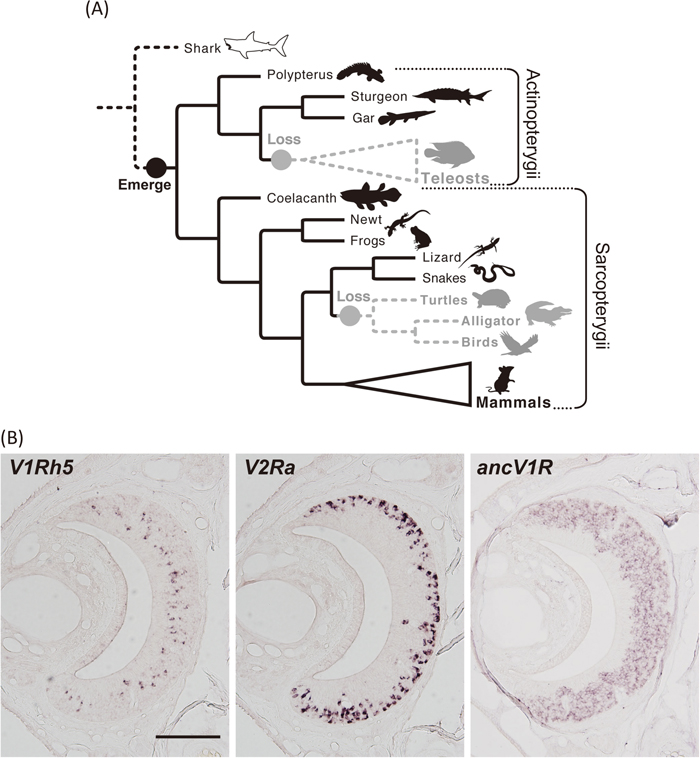
(A) Evolutionary scenario of ancV1R during vertebrate evolution. ancV1R emerged in the common ancestor of Sarcopterygii and Actinopterygii more than 400 million years ago and was lost in each of the common ancestors (gray circles) of teleost fishes as well as turtles, alligators and birds (gray dashed lines and silhouettes) by pseudogenization. The white silhouette and dashed black lines associated with sharks indicate that ancV1R has not yet emerged. Figure adapted from Suzuki et al. (2018). (B) Distinct pattern of expression between canonical VRs and ancV1R in the VNO. In situ hybridization on coronal sections of the VNO of C57BL/6 mice was performed using probes against canonical V1R (V1Rh5, left) and V2R (V2Ra, middle), and ancV1R (right). Note that canonical V1R and V2R show punctate expression in the apical and basal regions of the vomeronasal epithelium, respectively, according to the one neuron–one receptor rule. However, ancV1R shows broad expression in most vomeronasal sensory neurons, suggesting a new “one neuron–two receptors” (canonical V1R or V2R + ancV1R) rule. Scale bar indicates 200 μm.
In addition to the function of the VNO, ancV1R may shed light on its origin. Given that ancV1R is expressed in most VSNs, it can be used as an ideal molecular marker to explore the VNO in these groups. Traditionally, the VNO was believed to have originated in the common ancestor of tetrapods (Bertmar, 1981). However, a primordial VNO was later identified in the lungfish, implying an earlier origin (González et al., 2010; Nakamuta et al., 2012). To examine the possibility of a much earlier origin of the VNO, we need to focus on the basal lineages of ray-finned fishes (e.g., Polypterus, gars), which possess ancV1R even though morphologically apparent VNOs have not been identified in these ancient vertebrates.
In this review, I provide a summary and overview of the evolutionary dynamics of V1Rs in a broad range of vertebrates. Several recent findings allow us to more accurately guide further investigation of the evolution and function of V1Rs and their role in driving the diversification of vertebrates. Specifically, the evolution of f-V1Rs and t-V1Rs can be more precisely elucidated by exploring V1R sequences in basal lineages of ray-finned fishes, whose genomes are now becoming available. Clarifying the function of ancV1R is of primary importance in understanding the mechanism of pheromone detection, much of which remains to be elucidated. To test these novel discoveries in a diversity of non-model vertebrates within a hypothesis-driven framework, our group is also currently setting up the experimental system in well-established animal models to compare phenotypic differences (e.g., reproductive behavior, physiology) between wild type and ancV1R-knockout mice.
The illustration of the mouse vomeronasal organ in Fig. 1 was drawn by Ms. M. Nishimori at Tokyo Institute of Technology. The phylogenetic tree was constructed by Mr. Zicong Zhang at Tokyo Institute of Technology. The in situ hybridizations of mouse VNO sections in Fig. 4 were provided by Dr. J. Hirota at Tokyo Institute of Technology. This work was funded by JSPS KAKENHI (16H04820, 25440189), MEXT KAKENHI (221S0002), the Asahi Glass Foundation and the Hitachi Global Foundation to M. N.