2020 Volume 95 Issue 3 Pages 119-131
2020 Volume 95 Issue 3 Pages 119-131
A homolog of the bacterial ribosome rescue factor ArfB was identified in Arabidopsis thaliana. The factor, named AtArfB for Arabidopsis thaliana ArfB, showed ribosome rescue activity in both in vivo and in vitro assays based on the bacterial translation system. As has been shown for ArfB, the ribosome rescue activity of AtArfB was dependent on the GGQ motif, the crucial motif for the function of class I release factors and ArfB. The C-terminal region of AtArfB was also important for its function. The N-terminal region of AtArfB, which is absent in bacterial ArfB, functioned as a transit peptide for chloroplast targeting in tobacco cells. These results strongly suggest that AtArfB is a ribosome rescue factor that functions in chloroplasts.
Translation is terminated at the stop codon by the action of class I release factor(s). In the absence of in-frame stop codons, the ribosome continues translation until it reaches the 3′ end of mRNA and stalls there. Ribosome stalling at the 3′ end of mRNA lacking in-frame stop codons, referred to as non-stop mRNA hereafter, lowers the translation activity in the cell. Such a situation is deleterious to the cell and the stalled ribosome needs to be released from the non-stop mRNA. Both prokaryotic and eukaryotic cells are known to be equipped with several ribosome rescue systems to avoid the lethal effect of the accumulation of stalled ribosomes (Frischmeyer et al., 2002; van Hoof et al., 2002; Tsuboi et al., 2012; Saito et al., 2013; Abo and Chadani, 2014; Himeno et al., 2015). Bacterial cells such as Escherichia coli rescue the stalled ribosomes primarily by trans-translation, which is promoted by tmRNA (transfer-messenger RNA) and SmpB protein. In trans-translation, the tmRNA-SmpB complex first binds to the stalled ribosome as a tRNA mimic and allows the ribosome to resume translation by switching its template from non-stop mRNA to the mRNA-like domain of tmRNA. The ribosome then terminates translation at the stop codon on tmRNA. During trans-translation, the nascent polypeptide encoded by the non-stop mRNA receives a tmRNA-encoded short peptide sequence called the SsrA-tag at its C-terminus, and the SsrA-tagged polypeptide is degraded by cellular proteases such as ClpXP and Lon. The trans-translation system is widely distributed among bacteria, since the tmRNA-encoding ssrA gene exists in almost all bacterial genomes so far sequenced. In addition to this ubiquitous rescue system, some bacteria have alternative ribosome rescue systems. Escherichia coli cells are equipped with two alternative rescue systems, which are promoted by factors named ArfA and ArfB (alternative ribosome rescue factor A and B). ArfA was first found as a factor required for the viability of E. coli cells lacking ssrA (Chadani et al., 2010). The ArfA protein somehow recognizes the stalled ribosome, binds to its empty A-site, and recruits RF2, one of two class I release factors of E. coli, to catalyze hydrolysis of peptidyl-tRNA at the P-site (Chadani et al., 2012; Shimizu, 2012; Kurita et al., 2014). As a result, the stalled ribosome is released from the non-stop mRNA. ArfB, which was identified as a multicopy suppressor of the synthetic lethality of an arfA and ssrA double mutation, is a homolog of authentic class I release factors (Chadani et al., 2011). It has the so-called GGQ motif, which resides in domain 3 of class I release factors and is crucial for hydrolysis of peptidyl-tRNA within the ribosome P-site. On the other hand, ArfB lacks domains 2 and 4 of class I release factors, which are required for termination codon recognition (Kogure et al., 2014), and acts as a stop codon-independent RF. While ArfA is found in a narrow range of bacterial species, ArfB is widely distributed not only in bacteria but also in mitochondria (Handa et al., 2011). A human mitochondrial ArfB homolog, ICT1 (immature colon carcinoma transcript-1), was previously shown to have ribosome rescue activity and is required for mitochondrial function (Handa et al., 2010; Richter et al., 2010; Feaga et al., 2016).
Mitochondria and chloroplasts have their own translation systems, which more closely resemble bacterial than cytoplasmic systems. For example, the chloroplast has two class I release factors, whereas the cytoplasmic translation system employs only one, eRF1. Chloroplast RF1 of Arabidopsis thaliana, AtcpRF1 (At3g62910), whose mutation causes an albino phenotype, complements the lack of RF1 in E. coli cells (Motohashi et al., 2007). Arabidopsis thaliana also has chloroplast RF2, whose mutation causes a weak growth defect (Meurer et al., 2002). These observations strongly suggest that chloroplasts and bacteria share similar mechanisms in translation termination. If the ribosome rescue system is universally important, mitochondria and chloroplasts may also have their own ribosome rescue systems.
Here we found a homolog of E. coli arfB in the A. thaliana genome. It encodes a protein localizing in chloroplasts, and the protein showed ribosome rescue activity in the E. coli translation system.
Ds53-2850-1 line (tag2 in Fig. 1B) was identified from RIKEN Ds tagged lines (Ito et al., 2002) as having a disrupted At1g62850 gene. RIKEN Ac/Ds tagged lines were generated using the Nossen ecotype. T-DNA insertion knockout lines based on the Columbia ecotype (SAIL_622_D06 (tag3 in Fig. 1B) and WiscDsLox425H11 (tag1 in Fig. 1B)) with a disrupted At1g62850 gene were obtained from the Syngenta Arabidopsis Insertion Library (SAIL) collection and the WiscDsLox T-DNA collection at the Arabidopsis Biological Resource Center (Sessions et al., 2002; Woody et al., 2007). The genotypes at various loci were confirmed by PCR using the primers Ds53-2850-1-F (a) or RF-SAIL622D06-F (b) with RF-SAIL622D06-R (c) (Fig. 1B).

AtArfB. (A) Comparison of the sequences of AtArfB, ICT1 and ArfB. The GGQ motif and arginine residue analyzed in this study (R228 of AtArfB) are indicated by asterisks. Residues conserved among all three proteins or conserved in two out of three are indicated by black boxes or gray boxes, respectively. (B) Schematic drawing of AtArfB. Exons and introns are shown by rectangles and lines, respectively. Dark gray boxes show untranslated regions. The positions of insertion mutations are shown by triangles (tag 1–3). PCR primers used (Ds53-2850-1-F (a), RF-SAIL622D06-F (b), RF-SAIL622D06-R (c), 12455FwBam (d), At1g62850-GFP-F (e), At1g62850-GFP-R (f), At1g62850-nostop-R (g), 12455RvKpn (h); see Table 3 for sequences) are shown by arrows. (C) RT-PCR data showing the expression of AtArfB (upper panels) and TUBULIN4 (TUB, lower panels) in each atarfB mutant and in their wild type background ecotypes. Col: Columbia ecotype; tag1: WiscDsLox425H11; Nos: Nossen ecotype; tag2: Ds53-2850-1, tag3: SAIL_622_D06.
Escherichia coli strains used in this study are listed in Table 1. Cells were grown aerobically at 37 ℃ in LB medium (1% (w/v) NaCl, 0.5% (w/v) yeast extract, 1% (w/v) Peptone S) or on LB agar plates containing 1.5% (w/v) agar. Isopropyl-β-D-thiogalactopyranoside (IPTG, 100 μM), glucose (0.1%), arabinose (0.1%), ampicillin (Amp, 100 μg/ml), chloramphenicol (Cm, 50 μg/ml) and kanamycin (Km, 50 μg/ml) were added to the medium when required.
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| BL21 | lon ompT (E. coli B strain) | Laboratory stock |
| CH111 | W3110 arfA::FRT ssrA::FRT | Chadani et al., 2011 |
| CH551 | BL21 slyD::FRT | Chadani et al., 2012 |
| KNO101 | W3110 slyD::FRT | Laboratory stock |
| SN1071 | F−, thr-1, leuB6, thi-1, lacY1, galK2, | Nozaki and Niki, 2019 |
| ara-4, xyl-5, mtl-1, proA2, his-60, | ||
| argE3, rpsL31, tsx-33, supE44, | ||
| recB21, recC22, sbcA23, ∆hsdR::frt | ||
| W3110 | WT (E. coli K12 strain) | Laboratory stock |
Plasmids and primers used in this study are listed in Tables 2 and 3, respectively. The AtArfB ORF was PCR-amplified, using primers 12455FwBam and 12455RvKpn, from a cDNA clone obtained from RIKEN BRC. The amplified fragment was digested with BamHI and KpnI and cloned into the corresponding site of pQE80L to construct pMN101. Site-directed mutagenesis was performed by an inverse PCR-mediated procedure using PrimeSTAR Max DNA polymerase (Takara) and a primer pair designed to have overlapping sequences according to the supplier’s direction. Primer pairs 12455FwGAQ and 12455RvGAQ, 12455R228AFw and 12455R228ARv, 12455dC5Fw and 12455dC5Rv, and 12455dC10Fw and 12455dC10Rv were used to construct pMN102, pMN103, pMN104 and pMN105, respectively. iVEC mutagenesis (Nozaki and Niki, 2019) was employed to delete the portion corresponding to the N-terminal 89 amino acids of AtArfB from pMN101 to construct pMN101S. pMN102S, pMN103S, pMN104S and pMN105S were then constructed by inverse PCR using primer pairs 12455FwGAQ and 12455RvGAQ, 12455R228AFw and 12455R228ARv, 12455dC5Fw and 12455dC5Rv, and 12455dC10Fw and 12455dC10Rv, respectively, with pMN101S as a template.
| Plasmid | Characteristics | Source |
|---|---|---|
| pQE80L | His tag fusion protein expression vector pBR322 replicon, Ampr | Qiagen |
| pBAD33 | p15A-derived cloning vector carrying Para expression system, Cmr | Laboratory stock |
| pBAD33-ssrA | derivative of pBAD33 carrying ssrA | Laboratory stock |
| pCH102 | p15A derivative carrying His6-protein-expressing system, lacIq, Kmr | Laboratory stock |
| pCH336 | derivative of pCH102 expressing His6-CRP non-stop | Laboratory stock |
| pCH352 | derivative of pQE80L expressing His6-CRP-FLAG non-stop | Laboratory stock |
| pCH400 | derivative of pQE80L expressing His6-ArfB | Laboratory stock |
| pCH410 | derivative of pQE80L expressing His6-ArfB (GAQ) | Laboratory stock |
| pMN101 | derivative of pQE80L expressing His6-AtArfB | This study |
| pMN102 | derivative of pQE80L expressing His6-AtArfB (GAQ) | This study |
| pMN103 | derivative of pQE80L expressing His6-AtArfB (R228A) | This study |
| pMN104 | derivative of pQE80L expressing His6-AtArfB (dC5) | This study |
| pMN105 | derivative of pQE80L expressing His6-AtArfB (dC10) | This study |
| pMN101S | derivative of pQE80L expressing His6-AtArfB (without transit peptide sequence) | This study |
| pMN102S | derivative of pQE80L expressing His6-AtArfB (without transit peptide sequence, GAQ) | This study |
| pMN103S | derivative of pQE80L expressing His6-AtArfB (without transit peptide sequence, R228A) | This study |
| pMN104S | derivative of pQE80L expressing His6-AtArfB (without transit peptide sequence, dC5) | This study |
| pMN105S | derivative of pQE80L expressing His6-AtArfB (without transit peptide sequence, dC10) | This study |
| pUGW6 | empty vector, CaMV 35S promoter-sGFP | Provided by |
| Prof. T. Nakagawa | ||
| APG2 tp::sGFP | derivative of pBI121 expressing APG2 transit peptide region-sGFP | Laboratory stock |
| under CaMV 35S promoter | ||
| AtArfB tp::sGFP | derivative of pUGW5 vector (provided by Prof. Nakagawa) | This study |
| expressing AtArfB transit peptide region-sGFP under CaMV 35S promoter | ||
| AtArfB::sGFP | derivative of pUGW5 vector (provided by Prof. Nakagawa) | This study |
| expressing AtArfB coding sequence-sGFP under CaMV 35S promoter |
| 12455FwBam | AAAGGATCCATGGCGGCAATCAGAACGA |
| 12455RvKpn | GGGGGTACCTTAATCATCGTAACTACCG |
| 12455FwGAQ | CCTGGAGCCCAGAATGTGAACAAATTGA |
| 12455RvGAQ | ATTCTGGGCTCCAGGACCACCACTTCTA |
| 12455R228AFw | AAATCTGCGGCAAGAAGCCGCGGTAGTTACGA |
| 12455R228ARv | GCTTCTTGCCGCAGATTTCTTGTCTGA |
| 12455dC5Fw | AGCCGCTAAAGTTACGATGATTAAGGT |
| 12455dC5Rv | GTAACTTTAGCGGCTTCTTCTCGCAGAT |
| 12455dC10Fw | GAAATCTTAAAGAAGAAGCCGCGGTAGT |
| 12455dC10Rv | GCGGCTTCTTCTTTAAGATTTCTTGTCTGACAG |
| 12445dN89Fw | CCATCACCATCACTCCGCTGGCAATGAGCCTACT |
| 12445dN89Rv | CATTGCCAGCGGAGTGATGGTGATGGTGATGCGA |
| Fwd-352PURE | AAGGAGATATACCAATGAGAGGATCGCATCACCAT |
| Rev-non-stop-352PURE | TAAAAAAGCCCGCTCATTAGGCGGGCTGCTTTCTG |
| PUREsystem universal primer | GAAATTAATACGACTCACTATAGGGAGAC |
| CACAACGGTTTCCCTCTAGAAATAATT | |
| At1g62850-GFP-F | CACCATGGCGGCAATCAGAACG |
| At1g62850-GFP-R | GACTTTCCGATCACCACCGCTTCC |
| At1g62850-nostop-R | ATCATCGTAACTACCGCGGCTTCT |
| Ds53-2850-1-F | AGATTTCTGAGAGTTGTCCCGAGAT |
| RF-SAIL622D06-F | GGTCCTGGAGGCCAGAATGTGAAC |
| RF-SAIL622D06-R | TGGAGGTGGAACATATGAAGCCG |
| TUB4-F | TTGCTGTCTTCGTTTCCCTGG |
| TUB4-R | GAGGGTGCCATTGACAACATC |
KNO101 cells harboring pCH336, which expresses His6-crp-NST mRNA upon IPTG induction, were transformed with the plasmids pQE80L, pMN101S, pMN102S, pMN103S, pMN104S, pMN105S, pCH400 or pCH410. Transformants were grown in 5 ml LB medium containing Amp, IPTG and Km for 5 h at 37 ℃ to allow expression of both the ribosome rescue factor and His6-CRP-NST. The cells were then suspended in 1 ml of water. An equal volume of 2× SDS (sodium dodecyl sulfate) sample buffer (100 mM Tris-HCl (pH 6.8), 10% 2-mercaptoethanol, 4% SDS, 0.02% bromophenol blue, 20% glycerol) was added to the suspension and incubated for 2 min at 95 ℃. Samples were separated by 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and further analyzed by anti-His6 Western blotting as described previously (Chadani et al., 2011). Protein bands of interest were visualized by ChemiLumi-One (Nacalai Tesque) and detected by LAS 4000 mini (GE Healthcare).
Protein purificationCH551 cells harboring pMN101S (His-dN89-AtArfB), pMN102S (His-dN89-AtArfB-GAQ) or pCH400 (His-ArfB) were grown in 40 ml of LB medium containing Amp and IPTG for 4 h. The cells were then harvested, suspended in buffer NA (50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, 10 mM imidazole) and disrupted by sonication. After removing the debris, His-tagged proteins were purified from this crude extract using Ni-NTA resin (QIAGEN).
In vitro transcription–translation analyses to assess ribosome rescue activity of AtArfBA DNA template encoding CRP-NST was prepared by two-step PCR. First, the crp ORF carried on pCH352 was amplified using Fwd-352PURE and Rev-non-stop-352PURE as primers. Using the resulting DNA fragment carrying the crp-NST ORF, second-step PCR was performed using PUREsystem universal primer and Rev-non-stop-352PURE as primers. All PCR reactions were performed using PrimeSTAR Max DNA polymerase (Takara). The second-step PCR product was subjected to phenol/chloroform extraction followed by ethanol precipitation. DNA was dissolved in 10 mM Tris-HCl (pH 8.0) and used as a template at the concentration of 25 ng/ml for a reconstituted cell-free transcription–translation reaction in PUREfrex 1.0 (GeneFrontier). The reaction mixture was incubated for 1 h at 37 ℃ for protein synthesis. After the reaction was completed, purified ribosome rescue factor (final concentration, 5 μM) or puromycin (2 mM) was added and the mixture was incubated for another 20 min at 37 ℃. An equal volume of 2× SDS sample buffer was added to stop the reaction and further incubated for 2 min at 37 ℃. Samples were separated in 12% NuPAGE (Invitrogen) and further analyzed by anti-CRP Western blotting as described previously (Chadani et al., 2012).
Constructs for expressing fusion proteins of AtArfB and synthetic green fluorescent protein (sGFP)The coding region and predicted transit peptide region of an AtArfB (RAFL14-80-M23) full-length cDNA clone provided by RIKEN BRC was cloned into the TOPO vector (Gateway cloning system, Invitrogen) (Seki et al., 2002). The coding region of AtArfB and predicted transit peptide region were amplified using the primers At1g62850-GFP-F (primer e in Fig. 1B) and At1g62850-GFP-R (f) or At1g62850-GFP-F (e) and At1g62850-nostop-R (g) and transferred into pUGW5 vector (provided by Prof. T. Nakagawa) using the Gateway system (Invitrogen) so that each sequence of AtArfB was fused in-frame to the N-terminus of sGFP in the pUGW5-sGFP vector.
Observation of subcellular localization of AtArfBCaMV35Sp::AtArfB tp::sGFP, 35Sp::AtArfB::sGFP and 35Sp::APG2 tp::sGFP constructs were introduced into Nicotiana tabacum (SR1) by the particle bombardment PDS-1000 System (Bio-Rad). Transient expression of the AtArfB tp::sGFP, AtArfB::sGFP and APG2 tp::sGFP fusion proteins in N. tabacum was observed by confocal laser scanning microscopy (LSM 700, Carl Zeiss). Particle bombardment and observation of sGFP signals were carried out as described previously (Motohashi et al., 2001).
Plant growth conditionsSterilized seeds were sown on germination medium (Valvekens et al., 1988) and kept at 4 ℃ for three days before being moved into a growth chamber. Plants were grown with 130 μmol m−2 s−1 at 22 ℃ under long-day growth conditions (16 h light/8 h dark). In the strong light condition, plants were grown with 130 μmol m−2 s−1 at 22 ℃ under long-day growth conditions (16 h light/8 h dark) for two weeks and then with 290 μmol m−2 s−1 at 22 ℃. Other particular conditions were that plants were grown directly by sowing seeds in soil, and the low temperature condition. In the low temperature condition, plants were grown with 130 μmol m−2 s−1 at 22 ℃ under long-day growth conditions (16 h light/8 h dark) for three weeks and then with 20 μmol m−2 s−1 at 4 ℃.
RNA extraction and RT-PCRTotal RNA was isolated from seedlings and organs using an RNeasy Mini Kit (QIAGEN) following the manufacturer’s instructions, and treated with RNase-free DNase I. First-strand cDNA was synthesized from about 1 μg total RNA from samples using a PrimeScript first-strand cDNA synthesis kit and an oligo(dT) primer (Takara). PCR conditions comprised 30 cycles of 15 s at 95 ℃, 5 s at 55 ℃ and 1 min at 72 ℃, using 100 ng cDNA and the primers RF-SAIL622D06-F (b) and RF-SAIL622D06-R (c) (Fig. 1B). TUBULIN4 cDNA was used as an internal standard of gene expression and was amplified using the primers TUB4-F and TUB4-R. RT-PCR analyses were performed in three replicates.
Measurement of the leaf area of atarfB mutantsLeaf area was measured using ImageJ software (http://imagej.nih.gov/ij/). Three-week-old plants were photographed from above, and the image was opened on ImageJ software. After measurement of all individuals, the data were analyzed by Excel (Microsoft) and graphed. We measured the leaf area three times and used 25 mutant and wild type plants each.
A BLAST search revealed A. thaliana genes whose products are homologous to E. coli ArfB. Two of them were the previously reported chloroplast RF1 and RF2 homologs (Meurer et al., 2002; Motohashi et al., 2007). Among the remaining homologs, we picked one gene, At1g62850, for further investigation because its translation product showed the highest similarity to E. coli ArfB and its human mitochondrial counterpart ICT1 (Fig. 1A). At1g62850 is a single-copy gene in the A. thaliana genome (TAIR10 CDS data). Several knockout lines of At1g62850 are available from RIKEN BRC (http://epd.brc.riken.jp/en/) and ABRC (https://abrc.osu.edu/order-stocks). We picked three tag lines, WiscDsLox425H11 (tag1), Ds53-2850-1 (tag2) and SAIL_622D06 (tag3), and analyzed the expression of At1g62850 using RT-PCR. In all three tag lines, At1g62850 was not expressed (Fig. 1B and 1C).
Suppression of synthetic lethality of arfA ssrA double mutant E. coli cells by the A. thaliana ArfB homologAt1g62850 ORF was cloned into the widely used E. coli expression vector pQE80L so that the expression of its product could be induced in the presence of IPTG as an N-terminally His6-tagged protein in E. coli cells. The resulting plasmid pMN101 was introduced into the E. coli strain CH111/pBAD33-ssrA. CH111 lacks its chromosomal arfA and ssrA genes. As the arfA ssrA double mutation is synthetically lethal, CH111/pBAD33-ssrA can only grow in the presence of arabinose, which induces the expression of plasmid-borne ssrA. The transformant, CH111/pBAD33-ssrA + pMN101, was analyzed for its colony-forming ability on LB agar plates. If the At1g62850-encoded protein expressed from pMN101 has ribosome rescue activity, this transformant might be viable even in the absence of SsrA. As shown in Fig. 2A, the transformant cells formed colonies on a plate containing glucose, where the expression of ssrA is repressed. This indicates that the N-terminally His6-tagged At1g62850 gene product expressed from pMN101 showed ribosome rescue activity in the cell. Based on this result, we named At1g62850 AtArfB (A. thaliana ArfB) and its protein product AtArfB. We also refer to N-terminally His6-tagged At1g62850 protein as His-AtArfB hereafter.

Suppression of synthetic lethality of arfA ssrA double mutant E. coli cells by AtArfB variants. (A) pQE80L-based plasmids expressing AtArfB or its variants as N-terminally His-tagged fusion proteins were introduced into CH111/pBAD33-ssrA and grown on LB agar medium containing glucose overnight at 37 ℃. (B) pQE80L-based plasmids expressing dN89-AtArfB or its variant as N-terminally His-tagged fusion proteins were introduced into CH111/pBAD33-ssrA and grown on LB agar medium containing glucose overnight at 37 ℃. For (A) and (B), schematic drawings of AtArfB variants are illustrated beneath each panel. The indicated transformant was streaked on the area shown in panel (C).
Like E. coli ArfB, AtArfB has the GGQ motif, a typical and critical sequence motif for class I release factors and their homologs. Disruption of this motif abolishes the ribosome rescue activity of ArfB both in vivo and in vitro (Chadani et al., 2011). Structural analysis revealed that this motif is positioned very close to the ester bond between polypeptide and tRNA of the peptidyl-tRNA in the ribosome P-site (Gagnon et al., 2012). From these and other observations, the GGQ motif is thought to be directly involved in the hydrolysis of peptidyl-tRNA, which leads to subsequent release of the ribosome in the class I RF pathways as well as the ArfB pathway. AtArfB also has a C-terminal portion rich in basic amino acid residues, as commonly seen in ArfB homologs (Chadani et al., 2010). To assess the importance of these features, we constructed His-AtArfB variants and analyzed their activity to suppress the synthetic lethal phenotype of the arfA ssrA double mutation using CH111/pBAD33-ssrA as a tester strain as described above. As shown in Fig. 2A, substitution of the GGQ motif sequence to GAQ (GAQ mutation) abolished the colony-forming activity of the transformant, indicating that this motif plays a crucial role in ribosome rescue. Deletion of the C-terminal five amino acid residues (dC5 mutation) resulted in the formation of smaller colonies, suggesting the importance of this region. Upon deletion of another five amino acid residues (dC10 mutation) or more (data not shown), no colonies were formed. Arginine at position 228 (R228), the ninth residue from the C-terminus of AtArfB, is shared among ArfB (R132), AtArfB and ICT1 (R201) (Fig. 1A). Substitution of this arginine to alanine also abolished the colony-forming activity of the tester strain. These observations suggest the importance of the C-terminal portion, especially R228, in the ribosome rescue activity of AtArfB.
Sequence comparison revealed that AtArfB has an additional 89 amino acid residues at its N-terminus that are absent in ArfB (Fig. 1A). This sequence is predicted to be a transit peptide that targets AtArfB to chloroplasts, and its deletion (dN89 mutation) increased the accumulation of His-AtArfB when expressed in E. coli cells (Supplementary Fig. S1). As shown in Fig. 2B, His-AtArfB with the dN89 mutation (His-dN89-AtArfB) suppressed the synthetic lethal phenotype of the arfA ssrA double mutant, as His-AtArfB did. Interestingly, R228A or dC5 mutations introduced into His-dN89-AtArfB had no effect on colony size. Moreover, His-AtArfB-dN89-dC10 suppressed the lack of ribosome rescue activity to some extent, probably because the increased level of the protein compensated for the negative effect caused by these mutations. We thus reasoned that R228A, dC5 and dC10 mutations diminished, but did not abolish, the protein’s activity to suppress the synthetic lethal phenotype of the arfA ssrA double mutation. On the other hand, His-AtArfB-dN89-GAQ did not show any suppression activity, suggesting that the GGQ motif is indispensable for the activity of AtArfB.
Ribosome rescue activity of AtArfB in E. coli cellsTo examine whether AtArfB suppresses the synthetic lethal phenotype of the arfA ssrA double mutation of E. coli by rescuing a ribosome stalled at the 3′ end of non-stop mRNA, we expressed the model non-stop mRNA together with His-dN89-AtArfB. pCH336 carries an artificially manipulated crp gene designed specifically to assess the activity of ribosome rescue factors. It has a rho-independent transcriptional terminator sequence within its ORF and is expressed as a non-stop mRNA. Expression of this gene, which we call crp-NST, causes ribosome stalling that can be rescued by cellular tmRNA- and SmpB-driven trans-translation (Fig. 3A). Note that the host E. coli strain used here has wild type ssrA and smpB, which are required for trans-translation. As a result, the translation product His6-CRP-NST is SsrA-tagged at its C-terminus and degraded. When active ribosome rescue factor is overexpressed in these cells, however, some proportion of the stalled ribosomes will be rescued by this alternative factor, in which case the translation product will not receive an SsrA-tag. As a result, it escapes proteolysis and can be detected. To make the result clear, this model crp has the SsrA-tag coding sequence at its C-terminus. If transcription does not terminate at the terminator, translation of this crp gene will be terminated at the stop codon; however, this translation product has an SsrA-tag sequence at the C-terminus and will be degraded. Upon incubation with anti-His6 antibody, therefore, CRP-NST should be detectable only when active ribosome rescue factor is present in the cell. Using this system, we assessed the ribosome rescue activity of His-dN89-AtArfB (Fig. 3B). As expected, His6-CRP-NST was detected when His-dN89-AtArfB was expressed in the cell, in good agreement with the suppression analysis described above (Fig. 2B). The same His6-CRP-NST product was clearly detected when His-dN89-AtArfB-R228A or His-dN89-AtArfB-dC5 was expressed in the cell. Expression of His-dN89-AtArfB-dC10 gave a faint band for the CRP-NST product, suggesting that this construct has weaker ribosome rescue activity. His-dN89-AtArfB-GAQ showed no ribosome rescue activity in this assay system. We thus reasoned that the suppression of the synthetic lethality of arfA ssrA double mutant E. coli by His-dN89-AtArfB could be attributed to its ribosome rescue activity. His-AtArfB, which contained the possible N-terminal transit peptide and suppressed the synthetic lethality of the arfA ssrA double mutation, showed greatly reduced rescue activity in this assay, presumably reflecting the low level of expression (data not shown).
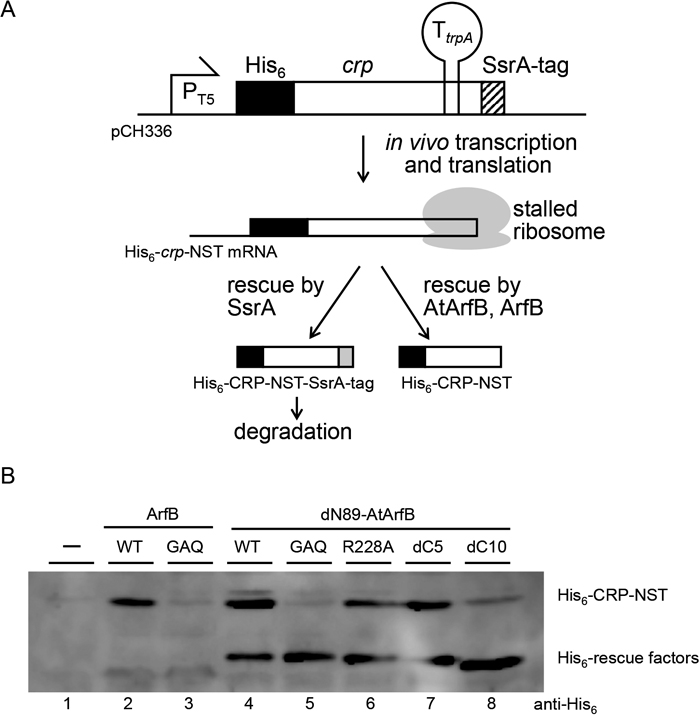
In vivo assay for ribosome rescue activity using artificially constructed non-stop mRNA. (A) Schematic drawing of the assay system. The crp gene carried on pCH336 has an extended ORF due to the disruption of the original termination codon. Because its termination codon is downstream of the trpA terminator (TtrpA), it will be transcribed as non-stop mRNA (His6-crp-NST). If the transcription does not terminate at TtrpA, full-length extended CRP, which is manipulated to have an SsrA-tag sequence attached at its C-terminus, will be produced and then soon degraded. As a result, only when the ribosome stalled at the 3′ end of His6-crp-NST mRNA is resolved by ribosome rescue systems other than trans-translation, the polypeptide encoded on the His6-crp-NST (His6-CRP-NST) will be detected. (B) KNO101 (W3110 ΔslyD::FRT) harboring pCH336 together with pQE80L (lane 1), pCH400 (lane 2), pCH410 (lane 3), pMN101S (lane 4), pMN102S (lane 5), pMN103S (lane 6), pMN104S (lane 7) and pMN105S (lane 8) were grown in LB (Amp, Km, IPTG) for 5 h at 37 ℃ and harvested. Total proteins were separated by 12% SDS-PAGE and further analyzed by anti-His6-tag Western blotting. Positions of His6-CRP-NST and rescue factors are shown on the right side of the panel.
To confirm the ribosome rescue activity of AtArfB, we also carried out in vitro translation analysis. crp-NST mRNA was translated in the PURE system, an E. coli-based reconstituted transcription–translation system. The ribosome is expected to stall at the 3′ end of crp-NST mRNA with peptidyl-tRNA in its P-site. If the ribosome is rescued, peptidyl-tRNA will be hydrolyzed. Ribosome rescue factor activity can thus be assessed by the disappearance of the peptidyl-tRNA, which can be detected by neutral-PAGE followed by Western blotting. The template DNA was expressed in the PURE system, and ribosome rescue factor was then added to allow ribosome rescue (Fig. 4). Puromycin, an antibiotic which resolves the peptidyl-tRNA by accepting the peptide portion of peptidyl-tRNA as a mimic of aminoacyl-tRNA, was also added as a control. When His-dN89-AtArfB, His-ArfB or puromycin was added, peptidyl-tRNA was not detected, whereas it was detected in the absence of any ribosome factors or in the presence of His-dN89-AtArfB-GAQ, which can neither suppress the synthetic lethality of arfA ssrA double mutant E. coli nor show ribosome rescue activity in vivo. From these results, we concluded that AtArfB has ribosome rescue activity in the E. coli system in a GGQ motif-dependent manner.
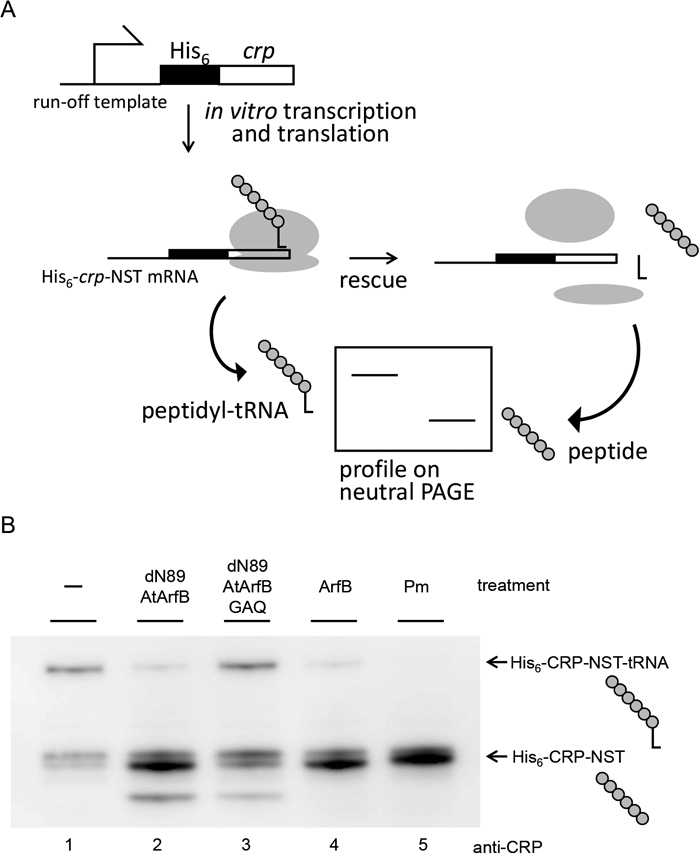
Ribosome rescue activity assayed in vitro. (A) Schematic drawing of the assay system. The artificial template DNA fragment used in this assay has a crp gene without in-frame stop codons so that transcription of this gene results in run-off crp-NST mRNA production. This fragment was expressed in the PURE system for 1 h at 37 ℃ to allow the ribosome to stall at the 3′ end of crp-NST mRNA. Purified ribosome rescue factor or puromycin was added to the reaction and the mixture was incubated for another 20 min at 37 ℃. The stalled ribosome contains peptidyl-tRNA, which is hydrolyzed to yield nascent polypeptide and tRNA upon ribosome rescue. Peptidyl-tRNA and polypeptide can be distinguished using neutral PAGE. Ribosomal 50S and 30S subunits are shown as large and small ovals, respectively. L-shaped tRNA is also illustrated. (B) Gel profile of the reaction products treated with AtArfB-dN89 (lane 2), AtArfB-dN89-GAQ (lane 3), ArfB (lane 4) or puromycin (Pm; lane 5), together with the reaction treated with no rescue factors (lane 1). The reactions were separated by 12% neutral SDS-PAGE (NuPAGE, Invitrogen) followed by anti-CRP Western blotting.
The N-terminal portion of AtArfB may be a transit peptide that targets this protein to the chloroplast. To test this, the coding sequence for the entire polypeptide or the possible transit peptide of AtArfB was cloned into the pUGW5-sGFP vector so that the resulting plasmid expresses sGFP fused with either AtArfB (AtArfB::sGFP) or the predicted transit peptide (AtArfB-tp::sGFP) from the CaMV (cauliflower mosaic virus) 35S promoter. We introduced the two constructs into N. tabacum (SR1) cells using the particle bombardment PDS-1000 System (Bio-Rad). As a control for transition to the chloroplast, the a construct expressing APG2-tp::sGFP (sGFP fused with the transit peptide of APG2) was also introduced into N. tabacum (SR1) cells; it is known that APG2 protein localizes to the chloroplast (Motohashi et al., 2001). Transient expression of AtArfB-tp::sGFP, AtArfB::sGFP and APG2-tp::sGFP in N. tabacum cells was observed by confocal laser scanning microscopy (LSM 700, Carl Zeiss), which clearly showed that AtArfB is localized in the chloroplast (Fig. 5C and 5D). From these subcellular localization analyses, we speculate that AtArfB is involved in the translation activity of photosynthetic proteins and influences photosynthetic activity.
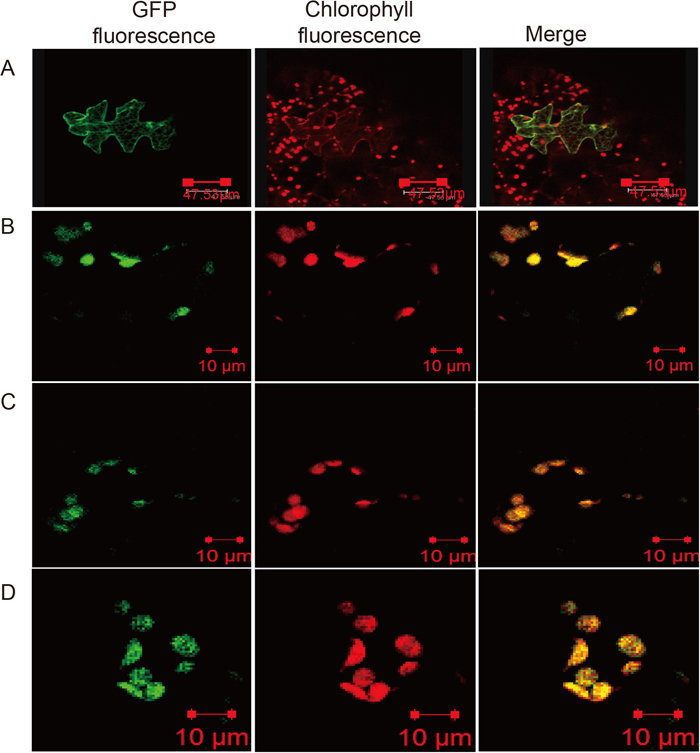
Intracellular localization of AtArfB in plants. The putative transit peptide region from AtArfB (N-terminal region shown in Fig. 1) and the ORF of AtArfB were cloned into pUGW5, consisting of sGFP under the control of the CaMV 35S promoter. AtArfB tp::sGFP and AtArfB::sGFP constructs were introduced into leaves of Nicotiana tabacum SR1 by particle bombardment. Cells expressing GFP were observed with a confocal laser scanning microscope. (A) pUGW6 (empty vector, CaMV 35S promoter::sGFP), sGFP without transit peptide was localized in cell membranes and the nucleus. (B) APG2 tp::sGFP (CaMV 35S promoter::APG2 transit peptide region::sGFP), APG2 transit peptide fused with GFP as a positive control was localized in chloroplasts. (C) AtArfB tp::sGFP (pUGW5 vector, CaMV 35S promoter::AtArfB transit peptide region::sGFP). (D) AtArfB::sGFP (pUGW5 vector, CaMV 35S promoter::AtArfB coding sequence::sGFP).
RT-PCR analysis showed that AtArfB was constitutively expressed in all organs (Supplementary Fig. S2). According to expression data in the Arabidopsis eFT Browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) (Winter et al., 2007), AtArfB is highly expressed under all conditions analyzed and no special condition was predicted to stimulate its expression. To better understand the regulation of AtArfB expression, we analyzed cis-elements in the 1,000-bp region preceding the start codon by PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and found 11 predicted light-responsive elements in the promoter region (data not shown).
Effect of AtArfB disruption on growth of A. thalianaAtArfB showed ribosome rescue activity in the bacterial translation system. Considering that the chloroplast translation system resembles the bacterial system, we questioned if AtArfB also acts as a ribosome rescue factor in the chloroplast. Unfortunately, the chloroplast translation system itself is very hard to analyze, and so we instead examined whether the disruption of AtArfB shows any phenotype. As described above, RT-PCR analysis revealed that AtArfB is not expressed in the tag lines WiscDsLox425H11 (tag1), Ds53-2850-1 (tag2) or SAIL_622D06 (tag3). Using these lines, we checked the effect of AtArfB disruption on germination, and on growth under strong light, at low temperature and in soil, but found no characteristic phenotype. However, WiscDsLox425H11 (tag1) and Ds53-2850-1 (tag2) lines displayed a 30% reduction in leaf area compared to the wild type (Fig. 6). Moreover, the three tag lines tended to have a significantly smaller plant size compared with the wild type (data not shown).
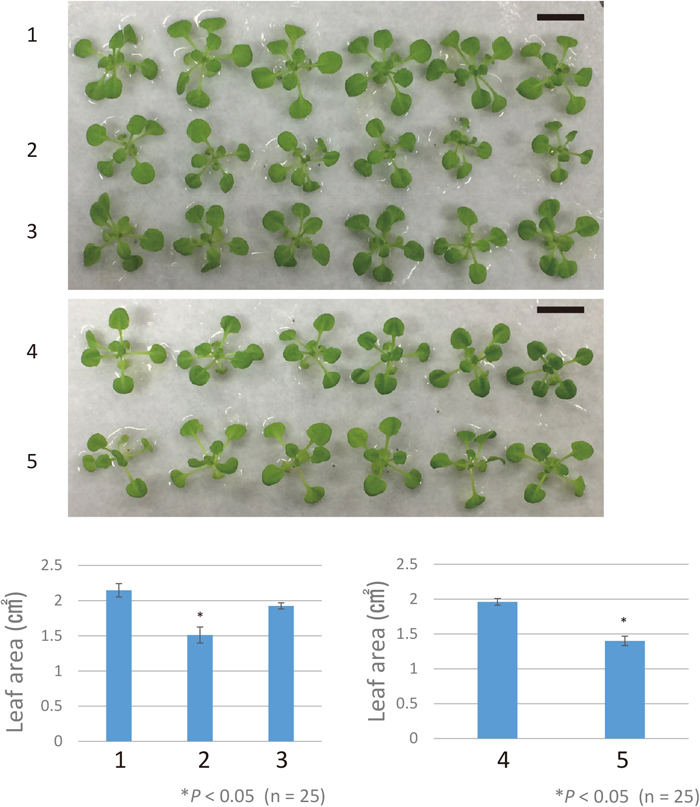
Leaf area of three tag lines. 1: Columbia; 2: WiscDsLox425H11 (tag1); 3: SAIL_622D06 (tag3); 4: Nossen; 5: Ds53-2850-1 (tag2). Bar: 1 cm. Mean leaf area is shown in the graphs beneath the photos. Error bars indicate standard deviation. Asterisks indicate a significant difference, using Student’s t-test, between WiscDsLox425H11 (2) and Columbia wild type (1), and between Ds53-2850-1 (5) and Nossen wild type (4).
AtArfB, an ArfB homolog in A. thaliana, was shown to have ribosome rescue activity when assayed in a bacterial translation system and also to be a chloroplast protein. AtArfB shares high sequence similarity with ArfB (26% identical) and ICT1 (37% identical). Like ArfB of E. coli, AtArfB rescues a ribosome stalled at the 3′ end of non-stop mRNA in a GGQ motif-dependent manner. The C-terminal portion of this protein, especially the R228 residue, is important for this activity. As this portion of ArfB has been shown to be important for the interaction with the ribosome (Chadani et al., 2011; Handa et al., 2011), the C-terminal portion of AtArfB may be similarly important. Considering that chloroplasts and mitochondria have their own translation system, which resembles the bacterial rather than the eukaryotic system, AtArfB probably functions as a ribosome rescue factor in the A. thaliana chloroplast. Despite the obvious ribosome rescue activity of AtArfB in a bacterial system, the AtArfB disruption itself did not show a clear phenotype. The leaf area of two tag lines having insertions upstream of the GGQ motif (tag1 and tag2) was smaller than wild type, but the tag lines showed no difference in Y(II) or Fv/Fm using a JUNIOR-PAM fluorometer (Walz) (data not shown).
One possible explanation for this result is that ribosome rescue activity is not as important in plant chloroplasts as in bacteria. If this were the case, however, we would need to explain why AtArfB is maintained in the A. thaliana genome and is expressed in various organs (Supplementary Fig. S2) and under various conditions. There are 11 light-responsive elements in the presumed promoter region of AtArfB. This suggests that AtArfB expression is responsive to light and involved in the translation of chloroplast proteins such as PrfB3 (a ribosomal release factor). PrfB3 functions in light- and stress-dependent regulation of stability of petB transcripts (Stoppel et al., 2011). Considering its resemblance to ArfB in terms of ribosome rescue activity in the bacterial system, namely GGQ motif dependency and the importance of the C-terminal portion, it is difficult to imagine that AtArfB has a function other than ribosome rescue. More plausible is the idea that there is an alternative ribosome rescue factor(s) in the chloroplast. There remain other arfB homologs in the A. thaliana genome that may also function in translation termination and/or ribosome rescue. A synthetic phenotype for AtArfB and other arfB homologs is under investigation. In any case, further analyses are required to understand the quality control system of translation termination in chloroplasts.
Exploring the ribosome rescue system in the chloroplast will shed light on the mechanism of how the translation system is maintained in the chloroplast, where biologically important molecules such as nucleic acids and proteins are exposed to various active molecules and radicals. Nucleic acids are especially sensitive to such hazardous agents, and various kinds of lesions may occur. To deal with such circumstances, preventing damage is of course necessary, but overcoming the translational deficiency caused by damaged nucleic acids, such as non-stop mRNAs, may also be important. Ribosome rescue may be one of the strategies that the energy-related organelles employ against the deleterious effect of reactive agents to sustain their normal function.
We gratefully acknowledge Prof. Tsuyoshi Nakagawa (Shimane University) for providing pUGW vectors to analyze the localization of proteins. RIKEN Arabidopsis tag line (Ds53-2850-1) was provided by RIKEN BRC which is participating in the National BioResource Project of the MEXT/AMED, Japan. This work was supported by the Ministry of Education, Culture, Sports, Science and Technology (Japan) [Grants-in-Aid for Scientific Research (No. 18K06350 to T. A.)].