Abstract
Understanding the control of anthocyanin biosynthesis is beneficial to genetic improvement for floral production in Dendrobium orchids. Full-length cDNA of CHS, CHI1, CHI2, F3H, DFR, ANS, F3'5'H, and FLS was isolated from Dendrobium hybrids with purple, peach, white and greenish white flowers. Analysis of the deduced amino acid sequences and gene expression levels of the eight genes suggested potential causes of color variation among the hybrids. Peach hybrid (SC) was likely due to changes in anthocyanin production from cyanidin to pelargonidin through mutations in F3'H, and the low color intensity was likely derived from the low expression levels of CHI1 and CHI2. In addition, white hybrid (RW) was likely caused by several mutations in F3H and/or high expression levels of FLS, an enzyme that converts color flavonoid intermediates into colorless flavonols. Simultaneous loss of F3H, DFR, and ANS expression observed in another white hybrid (JW) indicated that an alteration of anthocyanin regulatory controls was likely the cause of white coloration. Furthermore, analysis of hybrid mutants bearing pale and dark flowers demonstrated the influence of the expression of anthocyanin genes on the intensity of flower colors. Data obtained from this work could contribute to new strategies for future orchid breeding.
Introduction
Anthocyanins are a group of flavonoid glycosides constituting the major color pigments in flowers and fruit. Anthocyanins are synthesized along with flavonoid biosynthesis through a series of enzymatic reactions that convert chalcone into three major anthocyanidin types: cyanidin (red to magenta), pelargonidin (brick red to scarlet) and delphinidin (purple to violet) (see Tanaka et al., 2008, for reviews). Structural and regulatory genes are the key controls for the biosynthesis process. Spatial and temporal expression of the structural genes regulated through regulatory proteins dictates the production of anthocyanins in plants (Petroni and Tonelli, 2011).
Anthocyanin biosynthesis requires at least six enzymes including chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-β-hydroxylase (F3H), dihydroflavonol-4-reductase (DFR), anthocyanidin synthase (ANS) and flavonoid glycosyltransferase. Depending on the plant species, the product of F3H, dihydroflavonol, could be further substituted with hydroxyl groups by flavonoid 3'-hydroxylase (F3'H) and/or flavonoid 3',5'-β-hydroxylase (F3'5'H), generating three dihydroflavonol derivatives as intermediates for three branches of subsequent biosynthesis cascades. This leads to variations in pigment production via the three dihydroflavonols, namely, dihydroquercetin, dihydrokaempferol, and dihydromyricetin, which are then catalyzed by DFR and subsequently ANS, producing cyanidins, pelargonidins and delphinidins, respectively (Tanaka et al., 2008). Additionally, along with anthocyanin biosynthesis, flavone and flavonol are synthesized through the activity of flavone synthase (FNS) and flavonol synthase (FLS), respectively (Holton and Cornish, 1995; Martens and Mithöfer, 2005).
Many studies have demonstrated the effects of mutations in anthocyanin structural or regulatory genes on anthocyanin pigmentation. Loss-of-function mutations in CHS, CHI, F3H, DFR, and ANS normally cause a block in the biosynthesis, and plants harboring these mutations often produce white flowers or are colorless in tissues that usually contain color pigments (Britsch et al., 1992; Franken et al., 1991; Inagaki et al., 1996; Nakatsuka et al., 2005; Napoli et al., 1999). Studies of several yellow-flowered varieties of ornamental plants including Dianthus and Cyclamen showed that recessive mutations in CHI caused the accumulation of naringenin chalcone, resulting in yellow flowers (Forkmann and Dangelmayr, 1980; Miyajima et al., 1991). However, in some cases, CHI mutations did not result in complete disruption of anthocyanin production because some portion of the CHI substrate, naringenin chalcone, could be spontaneously catalyzed and proceed into the pathway (Forkmann and Dangelmayr, 1980; Miles and Main, 1985). Mutations in either F3'H or F3'5'H in many cases caused color alterations. Studies in roses, Dianthus and Chrysanthemum demonstrated that the lack of blue-purple in these plants was due to the loss of F3'5'H, which encodes the key enzyme responsible for delphinidin synthesis (Tanaka and Brugliera, 2006). F3'H switches anthocyanin biosynthesis to red-colored cyanidins and, in some varieties of Chrysanthemum in which F3'H is mutated, anthocyanin biosynthesis proceeds towards pelargonidin production resulting in orange flowers (Schwinn et al., 1993). Furthermore, certain types of mutation in DFR led to alterations in enzyme specificity towards its substrates. For example, while DFRs from many species such as Dianthus caryophyllus and Gerbera hybrida have broad specificity to the three types of dihydroflavonol, DFRs from petunia and Cymbidium cannot reduce dihydrokaempferol efficiently and, therefore, cannot produce pelargonidin-based color pigments (Forkmann and Rahnau, 1987; Helariutta et al., 1993; Johnson et al., 1999; Stich et al., 1992).
Dendrobium hybrids have been commercially distributed throughout the globe due to their elegant, colorful flowers with diverse shapes. Research has been conducted to understand the control of coloration in Dendrobium for improving flower production. However, knowledge regarding this issue is currently limited. Analysis of pigment compositions in 28 commercial Dendrobium species and hybrids has shown that cyanidins are the major pigment and pelargonidins are found in a few hybrids with peach or red flowers (Kuehnle et al., 1997). Previously, CHS and DFR genes were isolated and characterized to verify the difference of floral coloration controls in two Dendrobium hybrids producing purple and peach flowers. The analysis suggested that the substrate specificity of DFR was a possible cause of the color difference (Mudalige-Jayawickrama et al., 2005). Furthermore, CHS, DFR, and F3'5'H were recently isolated from D. moniliforme, and the analysis of F3'5'H expression suggested its role in floral coloration (Whang et al., 2011).
In this report, we aim to determine the coloration controls of four Dendrobium hybrids producing different flower colors including purple, peach, and two types of white. Full-length cDNA of CHS, CHI1, CHI2, F3H, DFR, ANS, F3'5'H, and FLS was isolated. Sequence and expression analyses revealed variations of deduced amino acid sequences and expression patterns of the eight genes in the four hybrids. Data obtained from this study were discussed on the basis of the different coloration in each hybrid. Furthermore, the analyses were extended to two selected Dendrobium mutants producing paler- and darker-colored flowers to address the possible causes of their color alterations.
Materials and Methods
Plant materials and RNA isolation
Four Dendrobium hybrids were obtained from local orchid farms in Thailand. These included a hybrid producing purple flowers named ‘Sonia Earsakul’ (SE), a peach-flowered hybrid called ‘Sirin classic’ (SC), and two white-flowered hybrids named ‘Suree white’ (RW) and ‘Jasmine white’ (JW). Two SE mutants that produce a paler color (L, ER6-329) and a deeper purple color (D, ER5-1129) on both petal and sepal were selected from the SE mutagenized population generated by acute gamma-irradiation of protocorm-like bodies at 30 Gy. The mutants were monitored for their consistency in color production in at least four consecutive flower generations. Four floral development stages, namely, early young bud (~0.5 × 1.5–2 cm: width × height), young bud (~1 × 2.5–3 cm), mature bud (~1.5 × 3.5–4 cm) and fully open flower stages, were collected for RNA isolation.
Total RNA was isolated from whole buds and flowers by a method described by Yu and Goh (2000). Briefly, buds or flowers were ground in liquid N2, suspended in the extraction buffer [50 mM CTAB, 40 mM Tris-HCl pH 7.5, 20 mM EDTA, 2 M NaCl, 1% (w/v) PVP-90 and 2% (v/v) β-mercaptoethanol] and incubated at 60°C for 15 min. The mixture was centrifuged at 7000 × g and 4°C for 15 min. Supernatant was mixed with an equal volume of chloroform:isoamylalcohol (24:1) and centrifuged at 7000 × g at 4°C for 15 min. This step was repeated twice. Supernatant was mixed with one-third volume of 10 M LiCl and kept at −20°C overnight for RNA precipitation. RNA was collected by centrifugation at 12000 × g at 4°C for 20 min and then washed with 2.5 M LiCl and 70% (v/v) ethanol in DEPC-treated water. RNA was dried and dissolved in DEPC-treated water. RNA was then subjected to DNase treatment (Promega, Medison, WI, USA), phenol:chloroform extraction and RNA precipitation. RNA was analyzed by agarose gel electrophoresis and quantified using Nanodrop (Thermo Scientific, Waltham, MA, USA).
5'- and 3'-RACEs and full-length cDNA cloning
Initially, partial sequences of CHS, CHI1, CHI2, F3H, DFR, ANS, F3'5'H, and FLS were amplified from cDNA of the SE hybrid, and these were used for priming sequences in 5'- and 3'-RACE reactions. Total RNA from buds and flowers of the SE hybrid was used for isolation of the 5' and 3' ends of the transcript of the eight anthocyanin biosynthesis genes using GeneRacer® Kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. 5' and 3' amplification reactions included 1 μL of GeneRacer-cDNA of the SE hybrid, 0.5 μM for either 5' or 3' RACE primers and gene-specific primers, 100 μM dNTPs, 1× Taq polymerase buffer and 0.5 units of Taq polymerase (NEB, Ipswich, MA, USA) in a total volume of 20 μL. PCR conditions were as follows: 2 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 50–60°C and 1 min at 72°C; and finally 72°C for 10 min. PCR products were resolved by electrophoresis in 1% (w/v) agarose gel, stained with ethidium bromide and visualized under UV light. DNA fragments were purified and cloned into the pGEM-T Easy Vector (Promega) for subsequent sequence analysis. 5' and 3' end sequences of each gene were used for primer design for full-length cDNA cloning.
For full-length cDNA cloning, cDNA of SE, SC, RW, JW, ER6-329, and ER5-1129 was synthesized using SuperScriptTM III Reverse Transcriptase (Invitrogen). Briefly, a reaction including 1 μg of total RNA, 2.5 μM oligo(dT)15–18, 0.5 mM dNTP and DEPC-treated water in a total volume of 13 μL was incubated at 65°C for 5 min before being cooled down on ice. Four microliters of 5× First-Strand buffer, 1 μL of DTT, 1 μL of RNaseOUTTM (Invitrogen) and 1 of μL SuperScriptTM III Reverse Transcriptase were added into the reaction and incubated at 50°C for 60 min and then at 70°C for 15 min. Full-length cDNA sequences of CHS, CHI, F3H, DFR, ANS, F3'5'H, and FLS were amplified from the cDNA with the primers listed in Table 1 using Easy-A® High-Fidelity PCR Cloning Enzyme (Stratagene, La Jolla, CA, USA) and subsequently sequenced. Nucleotide and deduced amino acid sequences were aligned using ClustalW version 3.
Quantitative real-time PCR analysis
cDNA was synthesized from total RNA obtained from young buds, mature buds and fully open flowers of SE, SC, RW and JW hybrids using SuperScriptTM III Reverse Transcriptase (Invitrogen). cDNA synthesis reaction was performed in the same manner as described in the full-length cDNA cloning section with the exception that 50 ng of random hexamers (Invitrogen) were used as replacement for 2.5 μM oligo(dT)15–18 primer. Quantitative real-time PCR was performed using 25-fold diluted cDNA samples. Each quantitative PCR reaction contained 7.5 μL of 2× SsoFastTM EvaGreen® Supermix (BioRad, Hercules, CA, USA), 2 μL of cDNA and 0.2 μM forward and reverse primers in a total volume of 15 μL (see Table 1 for primer sequences). Thermal cycling was performed on Eppendorf Mastercycler® ep Realplex Real-Time PCR Systems (Eppendorf, Hamburg, Germany) using a pre-heating step at 98°C for 30 s followed by a two-step cycle: 5 s at 98°C and 30 s at 60°C. 18S rRNA amplification was used as an internal standard. Data were analyzed using the ΔΔCt method with default parameters. Error bars represent the SD of three biological replicates and each was conducted in triplicate.
Semi-quantitative RT-PCR analysis
cDNA was synthesized from total RNA of the four flowering stages of SE, ER6-329 and ER5-1129 in the same manner as described for quantitative real-time PCR analysis. cDNA products were diluted 10-fold with dH2O, and semi-quantitative RT-PCR was performed in reactions containing 1 μL of cDNA, 0.5 μM for each primer, 100 μM dNTPs, 1× Taq polymerase buffer and 0.5 units of Taq polymerase (NEB) in a total volume of 20 μL. The reaction mixture was incubated in conditions of 2 min at 95°C; followed by 10–29 cycles of 30 s at 95°C, 30 s at the annealing temperature specific for each primer pair and 1 min at 72°C; and finally 10 min at 72°C (see Table 1 for primer sequences). 18S rRNA amplification was used as a reference. The analysis was performed in triplicate.
Results
Isolation and sequence analysis of full-length cDNA
Full-length cDNA of putative CHS, CHI, F3H, DFR, ANS, F3'5'H, and FLS was isolated from buds or flowers of Dendrobium SE hybrid. Two distinct putative CHI sequences, designated as CHI1 and CHI2, were isolated, whereas single amplicons were obtained from the other genes. This indicated that at least two CHI homologues are present in the SE hybrid genome. Alignments of deduced amino acid sequences of the isolated cDNAs with previously reported anthocyanin biosynthesis genes from other plant species showed high sequence similarity within each gene group (data not shown). The sequences were deposited in the NCBI database. Details of the eight full-length cDNA sequences regarding accession numbers, sequence lengths, deduced amino acids, the highest sequence identity match and their conserved domains are presented in Table 2.
Sequence analysis of the eight anthocyanin biosynthesis genes among four Dendrobium hybrids
To gain insight into the basis of color variations in flowers of Dendrobium hybrids, we examined nucleotide and deduced amino acid sequences of the eight genes from four hybrids that produce different flower colors. These included hybrids with flower colors ranging from dark purple (SE), peach (SC) and white (RW) to greenish white (JW) (Fig. 1). Nucleotide and amino acid sequence alignments showed a number of differences in nucleotide sequences that caused amino acid changes, a gene deletion and an immature translation termination in the eight genes from the four hybrids (Table 3). Open reading frames of the eight genes from SC, RW and JW hybrids were intact and similar to those from the SE hybrid in terms of both size and amino acid contents, except for CHI2 from RW and F3'5'H from JW. A single adenosine deletion at position 498 of the CHI2 coding sequence from RW was observed. This caused frame-shift mutation and consequently premature translational termination, which resulted in a shorter polypeptide product containing 156 rather than 209 amino acids. A 117-nucleotide deletion observed in F3'5'H from the JW hybrid resulted in a loss of 39 amino acids at position 7–45 compared with that of the SE hybrid. Furthermore, amino acid alterations located in the conserved region of each gene were also noted.

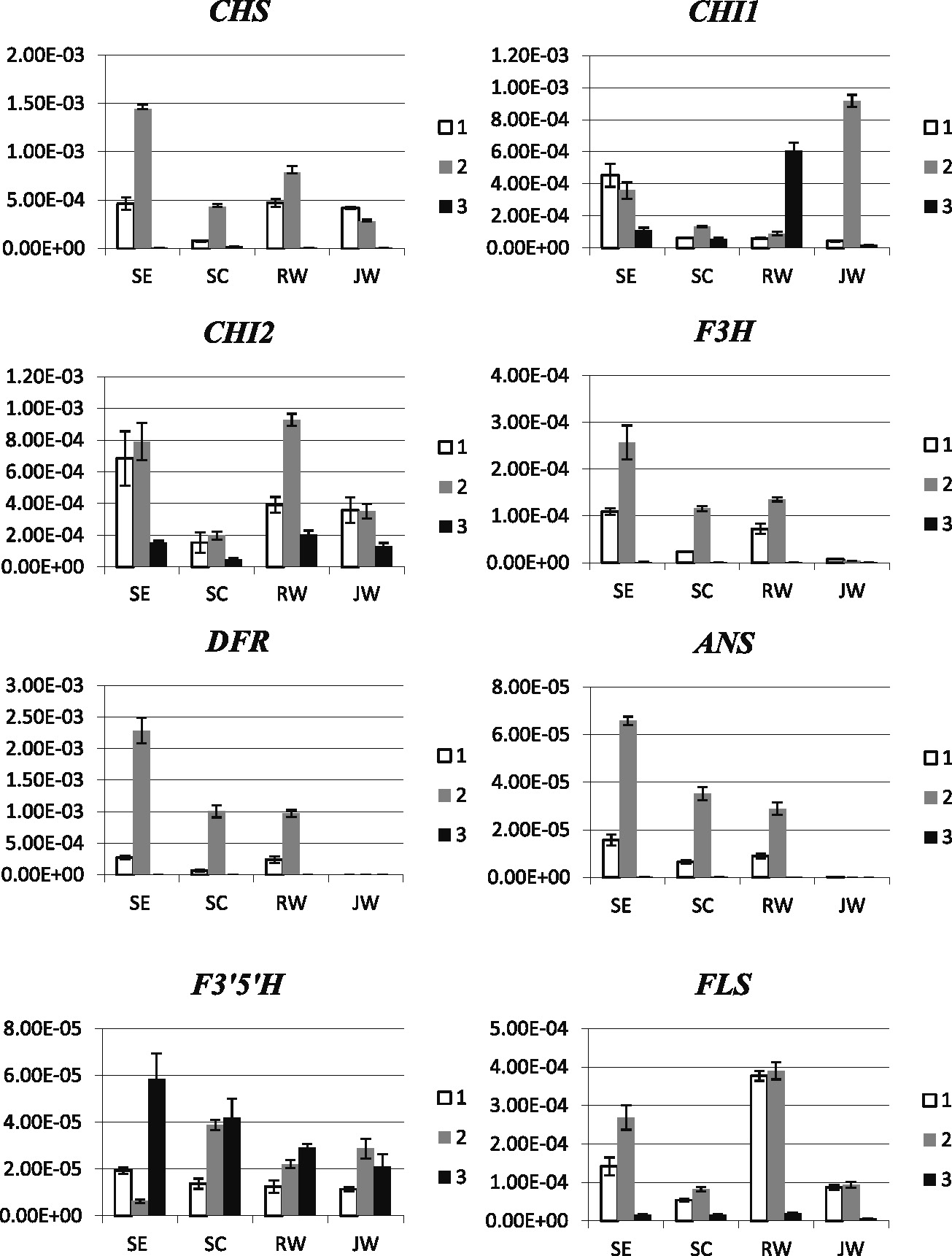
To examine the correlation between gene expression and flower colors, temporal expression of the eight genes in the four hybrids was analyzed using quantitative real-time PCR. The expression levels were monitored at three stages: young buds, mature buds, and anthesis. Variations in the expression levels of the eight genes were observed (Fig. 2). Generally, in all four hybrids, the expression of the anthocyanin biosynthesis genes was observed in young bud and gradually increased, reaching the highest levels in mature bud, before declining to very low levels at the anthesis stage, with the exception of F3'5'H, of which the expression remained high through to the anthesis stage. In the purple-flowered SE hybrid, the expression levels of CHS, CHI1, CHI2, F3H, DFR, ANS, and FLS were very high in young and mature buds compared with those from the other hybrids, before decreasing to very low or no expression in fully open flowers. In contrast, the expression levels of the eight genes in the SC hybrid were about one-third to one-half of those in the SE hybrid. The overall low levels of gene expression somehow reflected the fact that the color intensity of the SC hybrid flowers is far lower than that of the SE hybrid, regardless to the color. In white-flowered hybrids, distinct expression patterns were observed. We noticed that, in the JW hybrid, the expression of F3H, DFR, and ANS was almost undetectable and the expression of CHI1 was dramatically high compared with that of the other hybrids, whereas, in the RW hybrid, the expression level of FLS was distinctively high, about 5-fold higher than in the other hybrids, and CHI1 was expressed at a high level at the anthesis stage. Notably, the expression of CHI1 and that of CHI2 somehow compensated for each other. The expression of CHI2 was markedly high in young bud and mature bud stages but was reduced at the anthesis stage, whereas the expression of CHI1 was very low at the two early stages but abruptly increased in the anthesis stage.
We explored further the causes of the color changes in Dendrobium SE by examining two SE mutants that exhibit flowers with a paler color (L, ER6-329) and a darker color (D, ER5-1129) using sequence and expression analyses. Analysis of nucleotide sequences from eight anthocyanin biosynthesis genes showed no point mutation in both L and D mutants (data not shown). Semi-quantitative RT-PCR of CHS, F3H, F3'5'H, DFR, and FLS showed that, while the expression levels of the five genes in the wild type and the D mutant were similar, the expression levels in the L mutant were generally lower than those in both the wild type and the D mutant (Fig. 3). This indicated that the paler-colored flowers in the L mutant were the result of simultaneous reductions of gene expression in at least five genes involved in anthocyanin biosynthesis.
Discussion
The results from sequence alignments strongly suggest that the full-length cDNAs cloned from Dendrobium SE hybrid are indeed their corresponding anthocyanin biosynthesis genes. The alignment of CHS, CHI1, CHI2, DFR, and F3'5'H showed that sequences with the highest percent identity (74–100%) were from either Dendrobium or other orchid species. We noted that our CHS and DFR sequences were exact matches (100% identity) to previously described Den-CHS-4 and Den-DFR-1 isolated from Dendrobium Sw. (UH503) (Mudalige-Jayawickrama et al., 2005) and CHS from Dendrobium SE (Pitakdantham et al., 2010). Although there were no F3H, ANS, and FLS sequences available from orchids, our sequences were grouped very closely with those from monocots, with 71–95% identity.
Considering sequence alterations of the eight genes among the four hybrids, although some of the alterations could be anticipated, it was difficult to determine which specific amino acid changes would affect the protein functions and, therefore, alter the flower colors. According to the CHS alignment result, the alterations of CHS amino acid sequences observed in SC and JW hybrids were in variable regions and, thus, this is unlikely to affect the function of CHS. This is supported by the fact that the SC hybrid produced colored flowers. However, this does not preclude the possibility that this is the cause of white flowers in the JW hybrid. On the basis of a crystallography study of CHI protein structure (Jez et al., 2000), premature termination of the CHI2 in the RW hybrid, with more than 25% C-terminus polypeptide loss, is likely to result in functional disruption. Nevertheless, the presence of the CHI1 homolog is likely to compensate for this loss. We compared our deduced sequences to specific amino acid changes that have been previously reported to alter anthocyanin production, but no match was found. These changes included amino acid sequences of DFR from petunia, potato and Caryophyllales (De Jong et al., 2003; Johnson et al., 2001; Shimada et al., 2004), ANS from Caryophyllales, gentian and onions (Kim et al., 2005; Nakatsuka et al., 2005; Shimada et al., 2005), and F3'5'H and FLS from soybean (Takahashi et al., 2007, 2010). Nevertheless, it is worth noting that the amino acid alterations might potentially affect the flower colors. On the basis of the amino acid changes observed in the conserved regions of the coding proteins, these include changes in F3'5'H from the SC hybrid, F3H from the RW hybrid, and DFR and ANS from the JW hybrid.
The expression analysis showed that, in colored-flower hybrids including SE and SC, CHS, CHI, F3H, DFR, and ANS were generally expressed from young bud to mature bud stages. These expression data match with the flower colors as these five genes have been reported to be the key genes responsible for anthocyanin production (Mol et al., 1998). In all four hybrids, while the expression of the eight genes mostly occurred at the young and mature bud stages and dramatically declined to very low levels at the anthesis stage, the expression of F3'5'H was observed at elevated levels in the progression towards the anthesis stage. Despite the presence and expression of the F3'5'H gene, delphinidins have never been found to be the color pigments of Dendrobium species, and myricetin and syringetin 3',5'-hydroxylated flavonols were shown to represent F3'5'H enzymatic activity in Dendrobium (Kuehnle et al., 1997). Thus, our results suggest that the function of F3'5'H mostly occurs at late stages of flower development and this could contribute to the production of 3',5'-hydroxylated flavonols, which act as co-pigments for coloration in Dendrobium flowers.
Previous assessments determining the color pigments of the flowers of Dendrobium hybrids proposed a plausible cause of color changes from cyanidin type in lavender flowers of Dendrobium Sw. (UH503) to pelargonidin type in peach flowers of Dendrobium hybrid (K1224), which could be due to a mutation in F3'H, rather than in DFR (Mudalige-Jayawickrama et al., 2005). Such mutation resulted in the inefficient production of dihydroquercitin and led to more abundant dihydrokaempferol and, therefore, pelargonidins. This could be similar to our case in purple-flowered SE and peach-flowered SC hybrids. However, F3'H was not included in our study. Nonetheless, because the alterations of DFR sequences between SC and SE hybrids were neither in conserved regions nor in other positions that have been shown to affect its function, it could be anticipated that a mutation(s) in F3'H could be the cause of peach flowers in the SC hybrid.
Previous studies of anthocyanin contents suggested that the color intensity of Dendrobium flowers corresponded directly to pigment contents within the colored cells (Kuehnle et al., 1997). Considering the gene expression results, it is possible that the low color intensity in the SC hybrid could be because of the very low expression levels of CHI1 and CHI2. The reduction in CHI expression could, therefore, result in the low production of naringenin, downstream pigment production and color intensity, as a consequence.
Despite its white flowers, the expression profiles of the eight genes in the RW hybrid were similar and even higher for CHS, CHI1, and CHI2 than those in the SC hybrid. Two notable features were that the expression level of FLS was about 5-fold higher than those from the other hybrids and there was likely to be an interplay between the expression of CHI1 and CHI2. On the basis of our results, the production of white flowers in the RW hybrid could possibly be explained by either or both of the following causes. First, 6 amino acid alterations in the conserved regions of F3H of the RW hybrid compared with those of the SE hybrid suggest a high potential for F3H functional alteration. This could result in the blockage of subsequent anthocyanin biosynthesis and the loss of color pigments (Nakatsuka et al., 2005). Second, FLS catalyzes the production of colorless flavonols and competes with DFR in terms of anthocyanin production. Previous studies in white-flowered petunia containing high levels of flavonols demonstrated that the flower color could change from white to pink when either FLS was down-regulated or DFR was overexpressed (Davies et al., 2003). Giving that the CHI2 function was disrupted by premature termination and CHI1 expression was very low at early floral developmental stages, the level of the catalytic product of CHI could be very low in RW floral tissues. Thus, the white flowers of the RW hybrid could be the result of low expression levels of CHI1 and high expression levels of FLS, which cause the conversion of color flavonoid intermediates into colorless flavonols. To rule out these possibilities, further study of anthocyanin and flavonol compositions in RW flowers is required. If the RW hybrid exhibited white flowers because of CHI and/or F3H deficiencies, the accumulation of both anthocyanins and flavonols should be greatly reduced, whereas if this white flower phenotype is caused by the up-regulation of FLS, the accumulation of flavonols should be increased and that of anthocyanins should be reduced.
In the JW hybrid, F3H was hardly expressed throughout the flower stages, being about 30-fold less than those from the other hybrids. No expression was detected for DFR and ANS. These three genes are the key for anthocyanin pigment production (Martin and Gerats, 1993; Mol et al., 1998). A number of studies have shown that their expression had dramatic effects on color production. For example, loss or very low expression of either F3H or ANS was shown to be responsible for white flowers in Vanda hybrids and a gentian hybrid (Homoi), respectively (Junka et al., 2011; Nakatsuka et al., 2005). Simultaneous loss of expression in F3H, DFR, and ANS was also shown to be responsible for white coloration in both a gentian hybrid (Polano white) and a group of white-flowered Sim carnations (Mato et al., 2000; Nakatsuka et al., 2005). This simultaneous loss of expression is similar to our observation and we suggest that this would be the case for the white coloration in JW hybrid flowers.
Common regulatory factors controlling the expression of genes in the anthocyanin biosynthesis pathway are the key for colorations in flowers and other specific tissues (Davies et al., 1993). Analysis of white flowers in Sim carnation and gentians in comparison to their colored-flower counterparts suggested that mutations in regulatory proteins could result in the failure to induce a set of anthocyanin production genes during flower development (Mato et al., 2000; Nakatsuka et al., 2005). Thus, our results imply that, in the JW hybrid, F3H, DFR, and ANS were coordinately controlled, and the loss of expression of these genes was potentially due to a change in anthocyanin regulatory elements.
In maize, CHS, CHI, F3H, DFR, and ANS are transcriptionally regulated by three regulatory protein families known as MYB, bHLH, and WD40 (Petroni and Tonelli, 2011). In particular, in maize seeds, mutations of MYB-C1 or bHLH-R1 have been shown to cause a colorless phenotype, and a mutation in WD40-PAC1 was associated with reduced pigmentation (Dooner et al., 1991; Selinger and Chandler, 1999). Similarly, a recent report on Phalaenopsis showed that white coloration in the petals of P. amabillis resulted from the loss of anthocyanin-specific MYB transcripts, which subsequently caused the loss of DFR expression (Ma et al., 2009). From this, we suggest that the simultaneous reductions of gene expression of the five genes observed in L mutants was likely due to a mutation(s) in one of the regulatory genes in anthocyanin biosynthesis. Furthermore, a novel bHLH gene identified in maize (referred to as IN1) was shown to function as an inhibitor in anthocyanin biosynthesis, and in1 mutants exhibited very intense pigmentation (Burr et al., 1996). This may coincide with our observations and we suggest that the increase of color intensity in the flowers of D mutants might be because of a mutation in one of the bHLH inhibitors.
Data obtained from our study allowed postulations of factors affecting color variation in Dendrobium hybrids. Peach coloration in the SC hybrid is likely derived from the combination of changes in pigment production from cyanidin to pelargonidin through a mutation in F3'H and low levels of CHI1 and CHI2 expression. White coloration in the RW hybrid likely results from mutations in specific genes responsible for pigment production or increase in the expression of FLS that converts color pigments into colorless co-pigment molecules. Alternatively, in the JW hybrid, it is likely a result of the simultaneous loss of gene expression of a number of genes involved in the biosynthesis process. Furthermore, analysis of SE hybrid mutants bearing pale and dark flowers demonstrated that the expression levels of anthocyanin biosynthesis genes influence the intensity of color pigments in the flowers. Information obtained from these hybrids implying different approaches in flower coloration could benefit flower improvements by providing various strategies for genetic manipulations in Dendrobium and related orchids.
Acknowledgements
We are grateful to Supa Orchid Interlab for supplying protocorm-like bodies of Sonia Earsakul hybrid used in the mutagenesis study and providing support for orchid cultivation.
Literature Cited
- Britsch, L., B. Ruhnau-Brich and G. Forkmann. 1992. Molecular cloning, sequence analysis, and in vitro expression of flavanone 3 beta-hydroxylase from Petunia hybrida. J. Biol. Chem. 267: 5380–5387.
- Burr, F. A., B. Burr, B. E. Scheffler, M. Blewitt, U. Wienand and E. C. Matz. 1996. The maize repressor-like gene intensifier1 shares homology with the r1/b1 multigene family of transcription factors and exhibits missplicing. Plant Cell 8: 1249–1259.
- Davies, K. M., J. M. Bradley, K. E. Schwinn, K. R. Markham and E. Podivinsky. 1993. Flavonoid biosynthesis in flower petals of five lines of lisianthus (Eustoma grandiflorum Grise.). Plant Sci. 95: 67–77.
- Davies, K. M., K. E. Schwinn, S. C. Deroles, D. G. Manson, D. H. Lewis, S. J. Bloor and J. M. Bradley. 2003. Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica 131: 259–268.
- De Jong, W. S., D. M. De Jong, H. De Jong, J. Kalazich and M. Bodis. 2003. An allele of dihydroflavonol 4-reductase associated with the ability to produce red anthocyanin pigments in potato (Solanum tuberosum L.). Theor. Appl. Genet. 107: 1375–1383.
- Dooner, H. K., T. P. Robbins and R. A. Jorgensen. 1991. Genetic and developmental control of anthocyanin biosynthesis. Ann. Rev. Genet. 25: 173–199.
- Forkmann, G. and B. Dangelmayr. 1980. Genetic control of chalcone isomerase activityin flowers of Dianthus caryophyllus. Biochem. Genet. 18: 519–527.
- Forkmann, G. and B. Ruhnau. 1987. Distinct substrate specificity of dihydroflavonol 4-reductase from flowers of Petunia hybrida. Z. Naturforsch. 42C: 1146–1148.
- Franken, P., U. Niesbach-Klösgen, U. Weydemann, L. Marechal-Drouard, H. Saedler and U. Wienand. 1991. The duplicated chalcone synthase genes C2 and Whp (white pollen) of Zea mays are independently regulated; evidence for translational control of Whp expression by the anthocyanin intensifying gene in. EMBO J. 10: 2605–2612.
- Helariutta, Y., P. Elomaa, M. Kotilainen, P. Seppänen and T. H. Teeri. 1993. Cloning of cDNA coding for dihydroflavonol-4-reductase (DFR) and characterization of dfr expression in the corollas of Gerbera hybrida var. Regina (Compositae). Plant Mol. Biol. 22: 183–193.
- Holton, T. A. and E. C. Cornish. 1995. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7: 1071–1083.
- Inagaki, Y., Y. Hisatomi and S. Iida. 1996. Somatic mutations caused by excision of the transposable element, Tpn1, from the DFR gene for pigmentation in sub-epidermal layer of periclinally chimeric flowers of Japanese morning glory and their germinal transmission to their progeny. Theor. Appl. Genet. 92: 499–504.
- Jez, J. M., M. E. Bowman, R. A. Dixon and J. P. Noel. 2000. Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nature Struct. Mol. Biol. 7: 786–791.
- Johnson, E. T., S. Ryu, H. Yi, B. Shin, H. Cheong and G. Choi. 2001. Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase. Plant J. 25: 325–333.
- Johnson, E. T., H. Yi, B. Shin, B. J. Oh, H. Cheong and G. Choi. 1999. Cymbidium hybrida dihydroflavonol 4-reductase does not efficiently reduce dihydrokaempferol to produce orange pelargonidin-type anthocyanins. Plant J. 19: 81–85.
- Junka, N., S. Kanlayanarat, M. Buanong, S. Wongchaochant and C. Wongs-Aree. 2011. Analysis of anthocyanins and the expression patterns of genes involved in biosynthesis in two Vanda hybrids. Intl. J. Agr. Biol. 13: 873–880.
- Kim, S., R. Jones, K. S. Yoo and L. M. Pike. 2005. The L locus, one of complementary genes required for anthocyanin production in onions (Allium cepa), encodes anthocyanidin synthase. Theor. Appl. Genet. 111: 120–127.
- Kuehnle, A. R., D. H. Lewis, K. R. Markham, K. A. Mitchell, K. M. Davies and B. R. Jordan. 1997. Floral flavonoids and pH in Dendrobium orchid species and hybrids. Euphytica 95: 187–194.
- Ma, H., M. Pooler and R. Griesbach. 2009. Anthocyanin regulatory/structural gene expression in Phalaenopsis. J. Amer. Soc. Hort. Sci. 134: 88–96.
- Martens, S. and A. Mithöfer. 2005. Flavones and flavone synthases. Phytochemistry 66: 2399–2407.
- Martin, C. and T. Gerats. 1993. Control of pigment biosynthesis genes during petal development. Plant Cell 5: 1253–1264.
- Mato, M., T. Onozaki, Y. Ozeki, D. Higeta, Y. Itoh, Y. Yoshimoto, H. Ikeda, H. Yoshida and M. Shibata. 2000. Flavonoid biosynthesis in white-flowered Sim carnations Dianthus caryophyllus. Sci. Hortic. 84: 333–347.
- Miles, C. O. and L. Main. 1985. Kinetics and mechanism of the cyclisation of 2',6'-dihydroxy-4,4'-dimethoxy-chalcone; influence of the 6'-hydroxy group on the rate of cyclisation under neutral conditions. J. Chem. Soc., Perkin Trans. 2: 1639–1642.
- Miyajima, I., T. Maehara, T. Kage and K. Fujieda. 1991. Identification of the main agent causing yellow color of yellow-flowered cyclamen mutant. J. Japan. Soc. Hort. Sci. 60: 409–414.
- Mol, J., E. Grotewold and R. Koes. 1998. How genes paint flowers and seeds. Trends Plant Sci. 3: 212–217.
- Mudalige-Jayawickrama, R. G., M. M. Champagne, A. D. Hieber and A. R. Kuehnle. 2005. Cloning and characterization of two anthocyanin biosynthetic genes from Dendrobium orchid. J. Amer. Soc. Hort. Sci. 130: 611–618.
- Nakatsuka, T., M. Nishihara, K. Mishiba and S. Yamamura. 2005. Two different mutations are involved in the formation of white-flowered gentian plants. Plant Sci. 169: 949–958.
- Napoli, C. A., D. Fahy, H. Y. Wang and L. P. Taylor. 1999. White anther: a petunia mutant that abolishes pollen flavonol accumulation, induces male sterility, and is complemented by a chalcone synthase transgene. Plant Physiol. 120: 615–622.
- Petroni, K. and C. Tonelli. 2011. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 181: 219–229.
- Pitakdantham, W., T. Sutabutra, P. Chiemsombat and C. Pitaksutheepong. 2010. Isolation and characterization of chalcone synthase gene isolated from Dendrobium Sonia Earsakul. Pak. J. Biol. Sci. 13: 1000–1005.
- Schwinn, K. E., K. R. Markham and N. K. Giveno. 1993. Floral flavonoids and the potential for pelargonidin biosynthesis in commercial chrysanthemum cultivars. Phytochemistry 35: 145–150.
- Selinger, D. A. and V. L. Chandler. 1999. A mutation in the pale aleurone color1 gene identifies a novel regulator of the maize anthocyanin pathway. Plant Cell 11: 5–14.
- Shimada, S., Y. T. Inoue and M. Sakuta. 2005. Anthocyanidin synthase in non-anthocyanin-producing Caryophyllales species. Plant J. 44: 950–959.
- Shimada, S., K. Takahashi, Y. Sato and M. Sakuta. 2004. Dihydroflavonol 4-reductase cDNA from non-anthocyanin-producing species in the Caryophyllales. Plant Cell Physiol. 45: 1290–1298.
- Stich, K., T. Eidenberger, F. Wurst and G. Forkmann. 1992. Enzymatic conversion of dihydroflavonols to flavan-3,4-diols using flower extracts of Dianthus caryophyllus L. (carnation). Planta 187: 103–108.
- Takahashi, R., J. G. Dubouzet, H. Matsumura, K. Yasuda and T. Iwashina. 2010. A new allele of flower color gene W1 encoding flavonoid 3'5'-hydroxylase is responsible for light purple flowers in wild soybean Glycine soja. BMC Plant Biol. 10: 155. DOI: 10.1186/1471-2229-10-155. <http://www.biomedcentral.com/bmcplantbiol/>.
- Takahashi, R., S. M. Githiri, K. Hatayama, E. G. Dubouzet, N. Shimada, T. Aoki, S. Ayabe, T. Iwashina, K. Toda and H. Matsumura. 2007. A single-base deletion in soybean flavonol synthase gene is associated with magenta flower color. Plant Mol. Biol. 63: 125–135.
- Tanaka, Y. and F. Brugliera. 2006. Flower colour. p. 201–239. In: C. Ainsworth (ed.). Flowering and its manipulation. Blackwell, Oxford.
- Tanaka, Y., N. Sasaki and A. Ohmiya. 2008. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 54: 733–749.
- Whang, S. S., W. S. Um, I. J. Song, P. O. Lim, K. Choi, K. W. Park, K. W. Kang, M. S. Choi and J. C. Koo. 2011. Molecular analysis of anthocyanin biosynthetic genes and control of flower coloration by flavonoid 3',5'-hydroxylase (F3'5'H) in Dendrobium moniliforme. J. Plant Biol. 54: 209–218.
- Yu, H. and C. J. Goh. 2000. Identification and characterization of three orchid MADS-box genes of the AP1/AGL9 subfamily during floral transition. Plant Physiol. 123: 1325–1336.