2015 Volume 84 Issue 4 Pages 323-326
2015 Volume 84 Issue 4 Pages 323-326
An understanding of the genetic mechanism underlying capsaicinoid biosynthesis is important for breeding both pungent and non-pungent peppers. Although Capsicum is one of the earliest domesticated plant genera, only mutations in acyltransferase (Pun1) and putative aminotransferase (p-AMT) have been reported as genetic causes of loss of pungency. ‘No.3341’ (C. chinense) from Bolivia is a non-pungent cultivar. To determine the reasons underlying the non-pungency of ‘No.3341’, its expression levels and deduced amino acid sequences of Pun1 and p-AMT were analyzed. In ‘No.3341’, the expression levels and deduced amino acid sequences of both genes were normal compared with those of the pungent cultivar ‘Habanero’. Inheritance of the non-pungency was analyzed in F1 and F2 populations obtained by crossing ‘No.3341’ with ‘Habanero’. The segregation ratio indicated that the non-pungency of ‘No.3341’ is controlled by a single recessive gene. Moreover, since F1 populations obtained by crossing ‘No.3341’ with a non-pungent pepper, ‘NMCA30036’ harboring a mutation in Pun1, or with ‘No.2’ and ‘No.80’ harboring mutations in p-AMT, were all pungent, Pun1 and p-AMT could not account for the non-pungency of ‘No.3341’. It appears that a novel genetic mechanism is responsible for the loss of pungency in ‘No.3341’.
Capsicum, a member of the Solanaceae, originates from, and was first domesticated in South and Central America (Singh, 2007). On the basis of archeological evidence, Capsicum was already domesticated in 6000 B.P., making it one of the earliest domesticated plant genera (Perry et al., 2007). Capsicum was introduced to Europe at the end of the fifteenth century, after the first voyage of Christopher Columbus, and its use as a new horticultural crop rapidly spread across the Old World.
The pungency of chili pepper fruit is caused by a group of compounds known as capsaicinoids (Bennett and Kirby, 1968). These unique compounds are exclusively produced by the fruit of Capsicum (Andrews, 1984). Pungent peppers are a widely traded spice, perhaps the most commonly consumed spice in the world, making capsaicinoids one of the most commonly ingested plant secondary metabolites (Govindarajan, 1985; Hadacek, 2002). At the same time, non-pungent peppers are also valuable vegetable crops because they are concentrated sources of vitamins from the A, B, C, E, and K groups (Singh, 2007). For this reason, understanding the capsaicinoid biosynthetic pathway is important because both pungent and non-pungent peppers are valuable targets for vegetable crop improvement through plant breeding.
Given that the pungency of pepper fruit is one of its most important traits, numerous studies have been conducted on it. In C. annuum L., C. chinense Jacq., C. frutescens L., and C. chacoense Hunz., recessive mutations of acyltransferase (Pun1) and putative aminotransferase (p-AMT) have been reported to be the genetic causes of loss of pungency (Koeda et al., 2014; Lang et al., 2009; Stellari et al., 2010; Stewart et al., 2005, 2007; Tanaka et al., 2010a, b). Considering the long history of Capsicum as a crop, it is strange that only mutations in Pun1 and p-AMT have been used for breeding non-pungent peppers. We hypothesized that a pepper whose non-pungent property is attributable to a different genetic mechanism does exist. To validate this hypothesis, we screened Capsicum collections and identified a non-pungent pepper, ‘No.3341’ (C. chinense), as such a candidate in our preliminary research. In the present study, the inheritance of non-pungency in ‘No.3341’ and the association of Pun1 and p-AMT with its non-pungency were investigated.
Five C. chinense cultivars, ‘No.3341’, ‘No.2’, ‘No.80’, ‘NMCA30036’, and ‘Habanero’, were used in this study. ‘No.3341’ originates from Bolivia and has bell-shaped fruit. ‘No.2’ and ‘No.80’ are non-pungent cultivars carrying the recessive allele of putative aminotransferase (p-AMT; p-amtNo.2/p-amtNo.2, p-amtNo.80/p-amtNo.80) (Koeda et al., 2014), ‘NMCA30036’ is a non-pungent cultivar carrying the recessive allele of acyltransferase (Pun1; pun12/pun12) (Stewart et al., 2007), and ‘Habanero’ is a pungent cultivar. F1 and F2 populations were obtained by crossing ‘Habanero’ with ‘No.3341’ to determine the inheritance pattern of fruit pungency. Other F1 populations were prepared by crossing ‘No.2’ with ‘No.3341’, ‘No.80’ with ‘No.3341’, and ‘NMCA30036’ with ‘No.3341’. All plants were grown on Kyoto University experimental farm from March to October in 2012 and 2013. Segregation data were evaluated by chi-square test.
Phenotyping of fruit pungencyThe capsaicinoid contents of fruit were confirmed using high-performance liquid chromatography (HPLC). After the fruit had been freeze-dried, capsaicinoids were extracted and quantified according to the method described by Koeda et al. (2014). The capsaicinoid content was calculated as the sum of capsaicin and dihydrocapsaicin.
cDNA sequence analysis of Pun1 and p-AMTThe full-length cDNA sequences of Pun1 and p-AMT were determined for ‘No.3341’ and ‘Habanero’. Pepper fruit was harvested at 20 days after flowering and the placenta was separated for RNA extraction. Total RNA was extracted and reverse-transcribed according to the method described by Koeda et al. (2014). For RT-PCR, CaActin (AY572427) was used as a positive internal control. The full-length cDNA sequence of Pun1 was amplified using Pun1-F and Pun1-R primer sets (Koeda et al., 2014). The full-length cDNA sequence of p-AMT was amplified using F1 and R1481 primer sets (Tanaka et al., 2010b). PCR was performed using Blend taq (Toyobo, Osaka, Japan). For all PCR reactions, the reaction mixtures were initially denatured at 94°C for 2 min, followed by 35 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min, terminating with 3 min of extension at 72°C. Electrophoresis in 1.0% (w/v) agarose gel was performed with the amplified PCR products. For all of the treatments, RT-PCR was performed with three biological replicates using independently prepared total RNA and similar results were obtained. The full-length sequences of Pun1 and p-AMT amplified by RT-PCR were cloned into the pTaq-1 cloning vector (BioDynamics Laboratory, Tokyo, Japan). Nucleotide sequencing was performed in an ABI PRISM 3100 genetic analyzer with an ABI PRISM BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA).
Capsaicinoid accumulation was monitored using HPLC throughout fruit development in ‘Habanero’ and ‘No.3341’. Fruits of ‘Habanero’ and ‘No.3341’ were immature green at 10 days post-anthesis (dpa) and 20 dpa and were mature red at 30 dpa. In ‘Habanero’, capsaicinoids were not detected at 10 dpa but were detected at 20 dpa and increased untill 30 dpa (Fig. 1). In contrast, capsaicinoids were not detected in ‘No.3341’ throughout fruit development (Fig. 1).
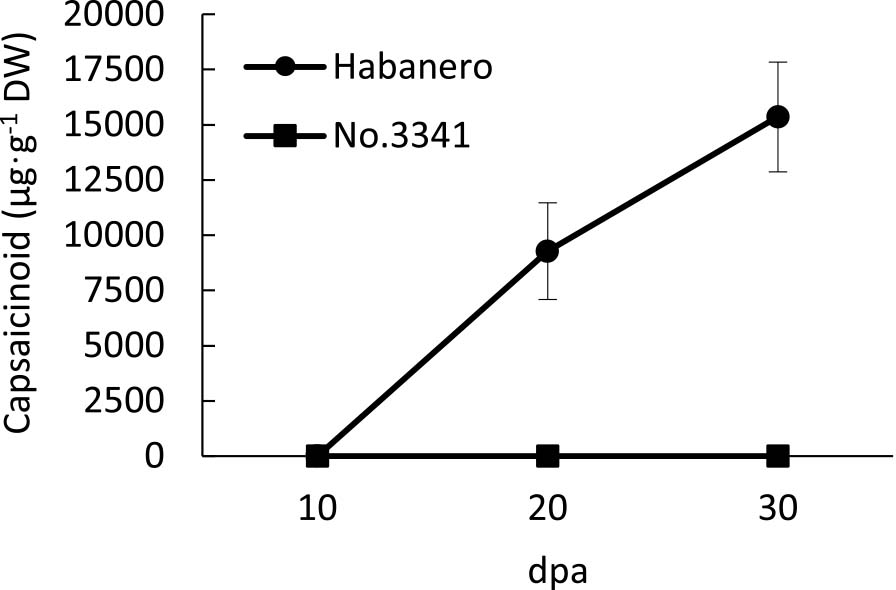
Capsaicinoid accumulation during the fruit development of ‘Habanero’ and ‘No.3341’. Capsaicinoid was not detected from ‘No.3341’. dpa: days post-anthesis. Error bars indicate standard deviation for the results of three plants.
We investigated the expression levels of Pun1 and p-AMT to elucidate the genetic basis of the non-pungent phenotype of ‘No.3341’. First, the expression of Pun1 was analyzed by RT-PCR. Pun1 fragments of 1.3 kbp from ‘Habanero’ and ‘No.3341’ were amplified (Fig. 2). This result indicates that at least ‘Habanero’ and ‘No.3341’ do not possess mutations such as a deletion spanning the promoter region that is observed in pun1 of C. annuum (Stewart et al., 2005). When cDNA sequences of Pun1 were compared for the two cultivars, the deduced amino acid sequences of Pun1 completely matched between ‘No.3341’ and ‘Habanero’ (data not shown). The expression level of p-AMT was also analyzed by RT-PCR. p-AMT fragments of 1.4 kbp from ‘Habanero’ and ‘No.3341’ were amplified (Fig. 2). Four recessive alleles of p-AMT have been reported from non-pungent C. chinense (Koeda et al., 2014; Tanaka et al., 2010b). All of them have small or large insertions in cDNA, which result in frameshift mutations. Such mutation was not observed in either ‘Habanero’ or ‘No.3341’ (Fig. 3). When cDNA sequences of p-AMT were compared for the two cultivars, it was found that amino acid residue 125 was leucine (L) in ‘No.3341’ but methionine (M) in ‘Habanero’ (Fig. 3). Given that residue 125 is not part of the functionally important PLP-binding domain and the deduced p-AMT amino acid sequences of pungent C. annuum (ADM13673.1) and C. frutescens (ADG65346.1) also coded L (Fig. 3), the difference between the deduced amino acid sequences of ‘No.3341’ and ‘Habanero’ was inferred not to be associated with the non-pungency of ‘No.3341’.

RT-PCR for full-length Pun1 and p-AMT in ‘Habanero’ and ‘No.3341’. Actin was used as a positive internal control.

Alignment of the deduced amino acid sequence of p-AMT from ‘Habanero’ and ‘No.3341’ with similar sequences of plant origin. p-AMT of ‘Habanero’ and ‘No.3341’ were aligned with p-AMT (C. annuum, ADM13673.1), p-AMT (C. frutescens, ADG65346.1), gamma aminobutyrate transaminase 2-like (S. lycopersicum, XP_004244777.1), and aminotransferase-like protein (A. thaliana, BAB03068.1). Underlined part indicates the PLP-binding domain. The 125 amino acid residues are shaded in gray.
To determine the genetic basis of the non-pungent phenotype, we crossed ‘No.3341’ with ‘Habanero’. F1 and F2 individuals that accumulated capsaicinoid at less than 92 μg·g−1 DW were categorized as non-pungent in this study. In several individuals, capsaicinoid was not detected. Because pungent individuals accumulated capsaicinoid between 1945 and 13820 μg·g−1 DW, pungent and non-pungent seemed to be a qualitative trait. The pungent and non-pungent phenotypes segregated 1:0 in F1 and 3:1 in F2 populations (Table 1), suggesting that the non-pungent phenotype of ‘No.3341’ is controlled by a single recessive gene. Moreover, because the F1 populations obtained by crossing ‘No.3341’ with ‘NMCA30036’, ‘No.80’, or ‘No.2’ were all pungent (Table 1), Pun1 and p-AMT could not account for the non-pungency of ‘No.3341’.

Phenotypic segregation of the non-pungent phenotype of ‘No.3341’.
Because pungency is one of the most important traits of pepper fruit as a crop, numerous studies have been conducted on it. In C. annuum, a single genetic source for non-pungency is suggested since the early identification in the 1500s of a widely distributed non-pungent pepper (Boswell, 1937), now known to carry a recessive allele of Pun1 (Stewart et al., 2005). Within C. annuum, most of the non-pungent cultivars carry the same single recessive allele of Pun1 (Stewart et al., 2005). Of late, two recessive alleles of p-AMT have been reported in two non-pungent cultivars (Lang et al., 2009; Tanaka et al., 2010a). In C. chinense, a single recessive allele of Pun1 and four recessive alleles of p-AMT have been reported to be the genetic basis of non-pungency (Koeda et al., 2014; Stewart et al., 2007; Tanaka et al., 2010b). Moreover, recessive mutations of Pun1 have been reported in C. frutescens and C. chacoense (Stellari et al., 2010). In the present study, we found that the loss of pungency in ‘No.3341’ is controlled by a single recessive gene that is neither Pun1 nor p-AMT. Our results indicate that a novel genetic mechanism is responsible for the loss of pungency in ‘No.3341’. Efforts to clone the gene responsible for the non-pungency of ‘No.3341’ are ongoing in our laboratory. Identifying this gene will reveal another approach for marker-assisted breeding of non-pungent peppers, and will shed light on the history of the breeding process of non-pungent cultivars.
We thank Toshio Sakakibara, Masaru Matsuda, and Koji Nishikawa (Experimental farm, Kyoto University) for their technical assistance in the field experiment, and Paul W. Bosland (NMSU Chile Pepper Institute) for providing ‘NMCA30036’.