2017 Volume 86 Issue 1 Pages 94-104
2017 Volume 86 Issue 1 Pages 94-104
The flowering response to monochromatic light and flowering-related genes underlying this response need to be characterized to efficiently use light-emitting diodes for lighting culture. The flowering response to far-red light has been well studied in long-day cut flowers, but there have been few studies investigating the response to blue light. Flowering and the expression of the G. paniculata homologs of the FLOWERING LOCUS T and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 genes (GpFT, GpSOC1) were not previously promoted in Gypsophila paniculata, an important long-day cut flower, ‘Bristol Fairy’ under long-day conditions with blue light. In the present study, we found that flowering was promoted in another G. paniculata ‘Million Star’, under long-day conditions with blue light, suggesting that there is variation in G. paniculata’s flowering response to blue light. Therefore, we analyzed the expression of GpFT and GpSOC1 in the ‘Million Star’. The expression of GpFT and GpSOC1 was promoted with flowering in ‘Million Star’ under long-day conditions with blue light in contrast to ‘Bristol Fairy’. We next analyzed the G. paniculata homologs (GpFKF1, GpGI) of FLAVIN-BINDING KELCH REPEAT F-BOX 1 and GIGANTEA genes, which participate in the flowering response to blue light. GpFKF1 and GpGI amino acid sequences were well conserved; gene expression showed a diurnal rhythm with different peaks under short-day and long-day conditions, as previously observed in Arabidopsis thaliana. GpFKF1 interacted with GpGI. There were no important differences in GpFKF1 or GpGI amino acid sequences between the two cultivars. Our results suggest that variation in the flowering response to blue light is associated with GpFT and GpSOC1, rather than GpFKF1 and GpGI.
Production of long-day cut flowers during autumn and winter is possible by long-day treatment to promote flowering. Although incandescent lamps have been used as a light source for long-day treatment, they have some disadvantages including high-energy consumption and short life. In contrast, light-emitting diodes (LEDs) are expected to reduce the environmental burden, owing to their low energy consumption and long life. Besides, because LEDs emit monochromatic light, lighting at the most effective wavelength can be used for long-day treatment. For these reasons, LEDs are receiving much attention as a new light source for long-day treatment.
The effects of monochromatic light quality on plant development should be investigated for the efficient use of LEDs in long-day treatment. Blue, red, and far-red lights are effective for plant flowering (Eskins, 1992), and their molecular mechanisms have been studied mainly in the model plant Arabidopsis thaliana (Andrés and Coupland, 2012; Shibuya and Kanayama, 2014).
FLOWERING LOCUS T (FT), which is known as florigen, with its main transcription factor CONSTANS (CO) plays a central role in the promotion of flowering in A. thaliana under long-day conditions (Corbesier et al., 2007; Wigge et al., 2005; Yoo et al., 2005). Recent studies have also shown that FT plays a key role in flowering in various horticultural plants (Nakajima et al., 2014; Xing et al., 2014). One of the blue light receptors regulating CO accumulation is CRYPTOCHROME 2 (CRY2) and the cry2 mutant shows late flowering under long-day conditions in A. thaliana (Guo et al., 1998). The other is FLAVIN-BINDING KELCH REPEAT F-BOX 1 (FKF1). FKF1 forms a complex with GIGANTEA (GI) in a blue-light-dependent manner and induces the degradation of CYCLING DOF FACTOR (CDF) 1 and its family proteins (CDFs), which are repressors of CO expression (Fornara et al., 2009; Imaizumi et al., 2003, 2005; Sawa et al., 2007). FKF1 also interacts with CO and stabilizes CO, and blue light enhances this interaction (Song et al., 2012). Furthermore, FKF1 removes CDF1, which also represses FT transcription, from the FT promoter (Song et al., 2012). FKF1 and GI thus play important roles in flowering induced by blue light. However, there are few studies on FKF1 and GI in long-day cut flowers.
Promotion of flowering by far-red or a low red:far-red ratio has been relatively well studied in terms of the flowering response of long-day cut flowers to light quality. For example, a low red:far-red ratio maintained using films that change light quality in daytime and red- or far-red-rich fluorescent lamps at night promote flowering in Matthiola incana (Yoshimura et al., 2002, 2006), and low and high red:far-red ratios promote and retard flowering under long-day conditions in Eustoma grandiflorum (Sato et al., 2009; Yamada et al., 2008a, b, 2009). Far-red alone and a low red:far-red ratio promote flowering under long-day conditions with LEDs in Gypsophila paniculata (Nishidate et al., 2012), and a low red:far-red ratio also promotes flowering in Campanula carpatica (Kristinsen, 1988). However, there are few studies on the flowering response to blue light in long-day cut flowers. Blue LEDs, in addition to far-red LEDs, represent a potential light source for lighting culture because blue light promotes flowering in the model plant A. thaliana (Eskins, 1992). Blue LEDs are also used with red LEDs in plant factories. Blue LED use has become increasingly common and widespread, and blue LEDs are inexpensive and also easier to obtain than far-red LEDs, which are not common. Using blue LEDs can thus help mitigate the costs associated with the use of LEDs to promote flowering.
For the efficient use of blue LEDs in lighting culture of long-day cut flowers, the flowering response to blue light and flowering-related genes underlying this response need to be characterized. We used Baby’s Breath (G. paniculata) in this study because it is often used in flower arrangements and is an important long-day cut flower (Kikuchi et al., 2000; Shillo and Halevy, 1982). G. paniculata is grown by lighting culture for year-round production and shows a clear flowering response to long-day treatment as a qualitative long-day plant (Hori et al., 2011; Nishidate et al., 2012). In a previous study (Hori et al., 2011), flowering and the expression of the G. paniculata homologs of the FT and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 genes (GpFT, GpSOC1) were not promoted in G. paniculata ‘Bristol Fairy’ under long-day conditions with blue light. In the present study, flowering was promoted in another G. paniculata ‘Million Star’, under long-day conditions with blue light, suggesting that there is variation in G. paniculata’s flowering response to blue light. We accordingly analyzed flowering-related genes to further investigate this variation.
G. paniculata ‘Million Star’ (Sumika Agrotech, Osaka, Japan) and ‘Bristol Fairy’ (Japan Agribio Company, Tokyo, Japan) were grown under short-day conditions with an 8-h photoperiod for approximately 4 weeks. Thereafter, 16-h light treatment was started following the method of Nishidate et al. (2012). Three lateral shoots of ‘Million Star’ plants with approximately 10 leaves were grown in a growth chamber at 22/18°C under natural light from 9:00 to 17:00 and under far-red or blue LEDs from 17:00 to 9:00 for observation of flowering, expression analysis of GpFT and GpSOC1, and sequencing of GpFKF1 and GpGI cDNAs. The plants were also grown in the dark from 17:00 to 9:00 as a control. Far-red and blue LEDs had peaks at 740 and 465 nm, respectively (Hori et al., 2011). LEDs of either color supplied 10–15 μmol·m−2·s−1, respectively.
For expression analysis of GpFKF1 and GpGI, ‘Bristol Fairy’ plants with approximately 14 leaves were used for long-day (16 h) treatment with incandescent light in an incubator at 22°C, and the plants were also grown under the short-day condition of an 8-h photoperiod.
Cloning of cDNA encoding GpFKF1 and GpGI from G. paniculata and comparison of their sequences between ‘Bristol Fairy’ and ‘Million Star’Total RNA was extracted from the leaves of ‘Bristol Fairy’ plants grown under long-day conditions as described previously (Hori et al., 2011). Extraction of total RNA, purification of poly (A)+ RNA, and reverse transcription were performed by the methods of Odanaka et al. (2002) and Kato et al. (2003). For cloning cDNA encoding GpFKF1, the primers for the PCR were 5'-CCA AGR GAY GTN GCW TCN ATY GG-3' and 5'-CAY GAN AGH GTR TGN CCC CA-3'. The primer sequences were based on the conserved sequences of FKF1 (AF216523) and its homologs in an ice plant (Mesembryanthemum crystallinum, AY371291) and other species. cDNAs for GpFKF1 containing the entire coding regions were cloned by 3'- and 5'-RACE (Moriguchi et al., 2006). For cloning cDNA encoding GpGI, the primers for the first PCR were 5'-CTC ATA WGA RCT RTA ACT CC-3' and 5'-TTC CTC AGC VGT TGA TYT KC-3' (Hecht et al., 2005). The PCR products were diluted and used for the second PCR using the primers 5'-ATG RAA ATW TTY GTN GCM ACH G-3' and 5'-GCT TSA TGR CTV ACR CAN ACR G-3'. The primer sequences for the second PCR were based on the conserved sequences of GI (AAF00092) and its homologs in the soybean (ABE81212), wheat (AAQ11738), and other species. cDNAs for GpGI containing the entire coding regions were cloned by 3'- and 5'-RACE (Moriguchi et al., 2006). cDNAs for GpFKF1 and GpGI were sequenced and then amino acid sequences deduced with GENETYX version 10 software (GENETYX CO., Tokyo, Japan).
Total RNA was extracted from the leaves of G. paniculata ‘Million Star’ plants using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) and used for reverse transcription with a QuantiTect Reverse Transcription Kit (Qiagen). The nucleotide sequences of the cDNAs of GpFKF1 and GpGI were then determined by direct sequencing of the PCR products amplified with the primers designed from the nucleotide sequences of cDNAs cloned from ‘Bristol Fairy’. Alignments were generated by GENETYX to compare the amino acid sequences of ‘Bristol Fairy’ and ‘Million Star’.
Gene expression analysisFor the expression analysis of GpFT and GpSOC1, ‘Million Star’ leaves were sampled between 16:00 and 16:30 every 2 weeks, beginning 2 weeks after the start of 16-h light treatment. Extraction of RNA and real-time PCR were performed following Hori et al. (2011). The following primer sets were designed using Primer 3 software: 5'-TGG GTA GGC AAA CTG TGT ACG-3' and 5'-GCT CCA CAT ATC CAA AGA CGA TAC-3' for GpFT1 (AB562502); 5'-CAG TGT ACC CTC CCG TGT G-3' and 5'-GGT GAT TTA AGG CGG CAA C-3' for GpFT2 (AB562503); and 5'-TAT GGC GTG ATG CCA AAG G-3' and 5'-CGC TAC TAA CGC GCT TAA TCC-3' for GpSOC1 (AB562504). The nucleotide sequences of the primer sets for GpACT are described above.
For the expression analysis of GpFKF1 and GpGI, ‘Bristol Fairy’ leaves were sampled every 3 h from zeitgeber time (ZT) 0 to ZT 21 at 8 weeks after the start of long-day treatment. Extraction of RNA, separation of genomic DNA from the RNA sample, reverse transcription, and real-time PCR were performed following Hori et al. (2011). The following primer sets were designed using Primer 3 software: 5'-GAA CCA TTT GAG GCC ATT TG-3' and 5'-TTT GTG CCT TCC TTT GCT G-3' for GpGI; 5'-AAC CGT GTT GGA GGA CAC TC-3' and 5'-AAG GAC GAC CGA ATC ACA AG-3' for GpFKF1; and 5'-GGC CGT TCT TTC ATT GTA CG-3' and 5'-TCC TGC TCG AAG TCA AGA GC-3' for GpACT (AB562505).
Yeast two-hybrid assayYeast two-hybrid assays were performed with the Matchmaker Gold Yeast Two-Hybrid System (Clontech, Palo Alto, CA, USA). GpFKF1 containing the light-oxygen-voltage (LOV) domain and the F-box domain (Asp57 to Pro376), AtFKF1 containing the LOV domain and the F-box domain (Gly18 to Thr333), the N-terminal region of GpGI2 (Met1 to Phe411 with 12 bp of the 5'-UTR), and the N-terminal region of AtGI (Met1 to Leu402) were amplified with the corresponding specific primer pairs (Table 1). The PCR products of GpFKF1 and AtFKF1 were cloned into the pGADT7 AD prey vector to obtain the GpFKF1AD and AtFKF1AD prey vector. The PCR products of GpGI2 and AtGI were cloned into the pGBKT7 DNA-BD bait vector to obtain the GpGI2BD and AtGIBD bait vector. These cloning steps were performed with an In-Fusion HD Cloning Kit (Clontech). A Yeastmaker Yeast Transformation System 2 (Clontech) was used for transformation of prey and bait vectors to Y2HGold competent cells (Y2HGold). The Y2HGold yeasts with prey were each plated on synthetic, defined (SD)/-Leu, SD/-Leu/X-α-Gal, and SD/-Leu/X-α-Gal/aureobasidin A (AbA) media to test self-activation of each prey vector. The Y2HGold yeasts with bait were each plated on SD/-Trp, SD/-Trp/X-α-Gal, and SD/-Trp/X-α-Gal/AbA media to test the self-activation of each bait vector. Then, each set of prey and bait vectors was co-transformed into Y2HGold.

Primers used for the amplification of cDNA inserted into each vector in yeast two-hybrid assays.
Quadruple dropout medium (QDO; SD/-Ade/-His/-Leu/-Trp) plates and QDO/AbA plates were used to assess the interaction between prey and bait proteins. The plates were incubated for 3 days at 30°C under blue LEDs (peak at 465 nm, 15 μmol·m−2·s−1) or in the dark. The set of pGADT7-T and pGBKT7-53 was used as the positive control and the set of empty prey pGADT7 and empty bait pGBKT7 was used as the negative control.
Cloning of the promoter regions of the GpFT1 geneThe 5'-upstream region of the GpFT1 genes was obtained using the Universal GenomeWalker 2.0 Kit (Clontech). Separate aliquots of genomic DNA were digested with four blunt-end restriction enzymes (DraI, EcoRV, PvuII, and StuI) and ligated to Genome Walker adaptors. The first PCR was performed using adaptor primer 1 (AP1) and gene-specific primers (GSP1). The second PCR was performed using a nested adapter primer 2 (AP2) and inner gene-specific primers (GSP2). The amplified PCR products were examined on an agarose gel, cloned into the pUC118 vector, and sequenced. The primers used are shown in Table 2.

Gene-specific primers (GSPs) used for genome walking to amplify the GpFT1 promoter regions.
Multiple comparisons by the Tukey–Kramer test were performed using Ekuseru-Toukei 2010 (Social Survey Research Information, Tokyo, Japan).
Because flowering is not promoted under long-day conditions with blue light in ‘Bristol Fairy’ (Hori et al., 2011), we investigated the effects of light quality on flowering in another cultivar, ‘Million Star’, for comparison. Flower budding began at 4 and 6 weeks after the start of 16-h light treatment with far-red or blue light, respectively (Fig. 1A). Flowering began at 8 weeks after the start of 16-h light treatment with far-red or blue light and the number of flowering shoots increased until 16 weeks (Fig. 1B). The numbers of budding shoots and flowering shoots per plant were very low under short-day conditions during the treatment. Flowering in ‘Million Star’ was thus promoted by 16-h light treatment not only with far-red light but also with blue light (Fig. 1).

Effects of light quality on flowering in G. paniculata ‘Million Star’. Changes in numbers of budding shoots (A) and flowering shoots (B) per plant during 16-h light treatment with blue light or far-red light (for reference). Plants were also grown under short-day conditions (control). Values indicate the mean of nine plants. Values with different letters at 16 weeks are significantly different at P < 0.05 by Tukey’s test.
In ‘Bristol Fairy’, flowering under long-day conditions with blue light is not promoted and the levels of GpFT1, GpFT2, and GpSOC1 mRNA are low (Hori et al., 2011). We accordingly investigated the effects of light quality on the expression of GpFT and GpSOC1 in ‘Million Star’, in which flowering was promoted under long-day conditions with blue light (Fig. 1). GpFT1, GpFT2, and GpSOC1 mRNA levels were low under short-day conditions during the treatment (Fig. 2A, B, C). The GpFT1 mRNA level was higher in advance of flower budding under long-day conditions with blue light than under short-day conditions (Fig. 2A). GpFT2 and GpSOC1 mRNA levels increased from 4 weeks to 8 weeks after the start of the treatment under long-day conditions with blue light (Fig. 2B, C). The expression of GpFT1, GpFT2, and GpSOC1 was analyzed under long-day conditions with far-red light, which promoted the flowering of ‘Million Star’, and the genes showed high expression compared to that under short-day conditions (Fig. 2A, B, C). GpFT1 showed high expression under long-day conditions with far-red light compared to that with blue light at 4 and 6 weeks after the start of the treatment.

Effect of light quality on the expression of GpFT1, GpFT2, and GpSOC1 during 16-h light treatment in G. paniculata ‘Million Star’. GpFT1, GpFT2, and GpSOC1 expression were analyzed by real-time PCR in ‘Million Star’ grown under long-day conditions with blue or far-red light (for reference) and under short-day conditions (control). Changes in the relative expression level are shown during 16-h light treatment. Error bars indicate the standard error of the mean of three biological replicates. The Y axis in GpFT2 is the same as that in GpFT1 to compare the two genes. Values with different letters in each week in advance of flowering are significantly different at P < 0.05 by Tukey’s test.
cDNAs containing the entire coding regions of the FKF1 and GI homologs, which were named GpFKF1 and GpGI, were cloned from G. paniculata ‘Bristol Fairy’ by RT-PCR with primers based on the conserved sequences and rapid amplification of cDNA ends (RACE). Because three different sequences of the FKF1 homologs were cloned and two of them showed extremely low expression, another sequence was used in this study, GpFKF1. LOV, F-box, and Kelch-repeat domains, as well as the amino acids essential for the function of FKF1, were well conserved in G. paniculata as shown by amino acid alignment (Han et al., 2004; Sawa et al., 2007; Song et al., 2012) (Fig. 3A). GpFKF1 was 69.9% and 86.4% identical to FKF1 from A. thaliana and the FKF1 homolog of Dianthus caryophyllus. GpFKF1 fell into the same cluster as the FKF1 homologs in a phylogenetic tree of deduced amino acid sequences of FKF1 and ZEITLUPE (ZTL) homologs (Fig. 3B).

Multiple alignment and phylogenetic tree of GpFKF1. (A) Amino acid sequence alignment of GpFKF1 (LC012310) and FKF1 homologs from various species. The asterisks indicate important amino acids for blue light reception and signaling of FKF1, and the gray line, dashed line, and dotted-line boxes indicate LOV domain, F-BOX, and KELCH-REPEAT, respectively. (B) A phylogenetic tree based on the amino acid sequences of GpFKF1, FKF1 homologs, and ZTL homologs. The tree was constructed by neighbor joining after sequence alignment using ClustalW2. Branch numbers refer to the percentages of replicates that support the branch using the bootstrap method (1000 replicates). The scale bar corresponds to 0.1 amino acid substitutions per residue. Species and accession numbers were as follows: FvFKF1 (Fragaria vesca, XM_004297384), GmFKF1 (Glycine max, XM_003530876), SlFKF1 (Solanum lycopersicum, XM_004228691), AtFKF1 (A. thaliana, NM_105475), DcFKF1 (Dianthus caryophyllus, Dca26663.1), McFKF1 (Mesembryanthemum crystallinum, AY371291), AtLKP2 (A. thaliana, NM_179652), DcZTL (D. caryophyllus, Dca11293.1), McZTL (M. crystallinum, AY371290), SlZTL (S. lycopersicum, XM_004242857), FvZTL (F. vesca, XM_004296401), GmZTL (G. max, NM_001248927), and AtZTL (A. thaliana, NM_125119). The accession numbers of DcFKF1 and DcZTL from D. caryophyllus were obtained from Carnation DB (Yagi et al., 2014; http://carnation.kazusa.or.jp/).
Two different sequences of the GI homolog were cloned from G. paniculata and named GpGI1 and GpGI2. The deduced amino acid sequence of GpGI1 was 98.6% identical to that of GpGI2, and 73% and 95.7% identical to those of GI from A. thaliana and the GI homolog from D. caryophyllus, respectively. The amino acids conserved in 6 sequences of GI and the GI homologs used for reference were also conserved in GpGI1 and GpGI2 (Fig. 4A), and GpGI1 and GpGI2 fell into the same cluster as GI from D. caryophyllus in a phylogenetic tree (Fig. 4B).

Multiple alignment and phylogenetic tree of GpGI. (A) Amino acid sequence alignment of GpGI1 (LC012311) and GpGI2 (LC012312) from ‘Bristol Fairy’ and GI homologs from various species. The asterisks indicate differing sequence for ‘Bristol Fairy’ and ‘Million Star’ in GpGI1, and the pairs of amino acid in ‘Bristol Fairy’/amino acid in ‘Million Star’ were Ala317/Ser, Ile694/Val, Ala698/Thr, Asp828/Asn, Met844/Thr, Glu847/Lys, and Ala981/Val. (B) A phylogenetic tree based on the amino acid sequences of GpGI and GI homologs from various species. The tree was constructed by neighbor joining after sequence alignment with ClustalW2. Branch numbers refer to the percentage of replicates that support the branch using the bootstrap method (1000 replicates). The scale bar corresponds to 0.1 amino acid substitutions per residue. Species and accession numbers were as follows: FvGI (F. vesca, XM_004290435), GmGI (G. max, AB554222), AtGI (A. thaliana, AF105064), DcGI (D. caryophyllus, Dca29500.1 in carnation DB), OsGI (Oryza sativa, NM_001048755), and ZmGI (Zea mays, EU302138).
The effect of long-day treatment on the expression of GpFKF1 and GpGI was analyzed in ‘Bristol Fairy’. The primers used in real-time PCR amplified both GpGI1 and GpGI2 cDNAs. The levels of GpFKF1 and GpGI mRNA showed a clear diurnal rhythm and their peaks differed between the long-day and short-day conditions (Fig. 5). GpFKF1 and GpGI mRNA levels showed peaks around dusk under the short-day condition and in the latter half of a light period under the long-day condition.

Changes in expression of GpFKF1 and GpGI in G. paniculata ‘Bristol Fairy’ during a day/night cycle. GpFKF1 and GpGI expression were analyzed by real-time PCR in G. paniculata ‘Bristol Fairy’ grown under short-day (A) and long-day conditions (B), and changes in the relative expression level are shown. Error bars indicate the standard error of the mean of three biological replicates. White and black bars represent white light and dark periods.
Interaction between the N-terminal region of GpGI and the LOV and F-box domains of GpFKF1 was analyzed by yeast two-hybrid assays with cDNAs cloned from ‘Bristol Fairy’ (Fig. 6). The sequence from GpGI2 cDNA was used for the assays because the amino acid sequences of GpGI1 and GpGI2 were almost identical and there was no difference in important amino acids between them. GpFKF1 interacted with GpGI2, and GpFKF1 and GpGI2 interacted with AtGI and AtFKF1, respectively. The same results were obtained in assays in the dark and under blue light.

Interaction between GpFKF1 and GpGI2 proteins in yeast. Left and right images show the results on a QDO (SD/–Ade/–His/–Leu/–Trp) plate and a QDO + AbA plate, respectively. Colonies that grew on a QDO plate contained both prey and bait plasmids, which included LEU2 and TRP1, respectively, and also expressed proteins that interacted with each other to activate HIS3 and ADE2 through two types of Gal4-responsive promoters. In addition, AUR1-C was activated by interactions of two proteins through another type of Gal4-responsive promoter, and its products conferred expression of AbAr (resistance to the antibiotic AbA). Experiments were performed in the dark or under blue light.
We compared the amino acid sequences of GpFKF1 and GpGI of ‘Bristol Fairy’ and ‘Million Star’ because the two cultivars showed a different flowering response to long-day conditions with blue light. The amino acid sequences of GpFKF1 and GpGI2 were identical between the two cultivars, but GpGI1 had some different amino acids (Fig. 4A). The differences found in GpGI1 between the two cultivars were inferred not to be essential for function because the differing amino acids were not in conserved regions or were substituted by similar amino acids in conserved regions.
Sequence comparison of the GpFT1 promoter region between ‘Bristol Fairy’ and ‘Million Star’Sequences of the GpFT1 promoter region were compared between the two cultivars because the GpFT1 mRNA level increased in advance of flower budding under long-day conditions with blue light in ‘Million Star’. There were several deletions and nucleotide substitutions and, in particular, the promoter region from ‘Bristol Fairy’ contained a large deletion corresponding to −1080 bp to −1382 bp upstream of the start codon of GpFT1 in ‘Million Star’ (Fig. 7). The deleted region contained an E-box (CANNTG) and a TCTAG sequence, which are reportedly associated with the response to blue light through the CRY pathway (Liu et al., 2013; Shaikhali et al., 2012).
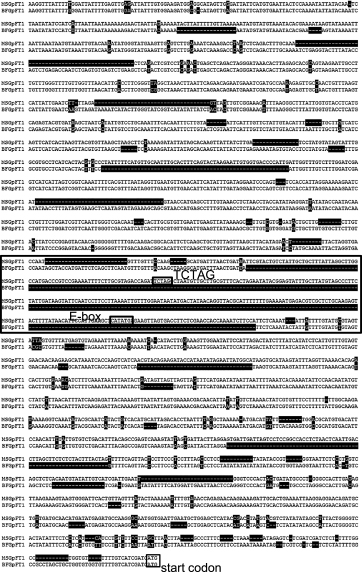
Nucleotide sequence alignment of the GpFT1 promoter regions from ‘Bristol Fairy’ (BF) and ‘Million Star’ (MS). The boxed region contains the longest deletion in BF, in which flowering was not induced under long-day conditions with blue light (Hori et al., 2011). An E-box and TCTAG motif, which may be important in blue light-dependent FT expression (Liu et al., 2013; Shaikhali et al., 2012), were found in this region from MS.
In our previous study (Hori et al., 2011), flowering was not promoted under long-day conditions with blue light in G. paniculata ‘Bristol Fairy’ despite the promotion of flowering by blue light in A. thaliana. We accordingly investigated the effect of blue light on flowering in other cultivars and found that flowering of ‘Million Star’ is promoted under long-day conditions with blue light. Our results revealed that there is variation in G. paniculata’s flowering response to blue light. Although the expression of GpFT and GpSOC1 in ‘Bristol Fairy’ is induced under long-day conditions with white light, their transcripts are low under long-day conditions with blue light, which does not promote flowering (Hori et al., 2011). In contrast, in the present study, the expression of those genes was induced under long-day conditions with blue light in ‘Million Star’. In particular, GpFT1 expression was higher in advance of flower budding under long-day conditions with blue light than under short-day conditions in ‘Million Star’. Under long-day conditions with far-red light, flowering is promoted in ‘Bristol Fairy’ with the expression of GpSOC1, whereas the expression of GpFT is not induced (Hori et al., 2011). Flowering is not promoted under long-day conditions with blue light in the ft mutant of A. thaliana unlike the response in the wild type, although flowering is promoted under long-day conditions with far-red light in the ft mutant and the wild type (Hori et al., 2011). This difference in the response to light quality between the ft mutant and the wild type in A. thaliana seems to have commonalities with the difference between ‘Bristol Fairy’ and ‘Million Star’ in G. paniculata.
We analyzed the G. paniculata homologs of FKF1 and GI, which act upstream of FT in a blue light-dependent manner in A. thaliana (Sawa et al., 2007; Song et al., 2012), because these genes may play an important role in G. paniculata’s flowering response to blue light. In addition, there is a lack of information on the flowering response to blue light in long-day cut flowers and there is, to our knowledge, no report of FKF1 and GI in cut flowers. The amino acid sequences of GpFKF1, GpGI1, and GpGI2 were found to be well conserved in a sequence comparison with the proteins from A. thaliana and other species. They were classified into FKF1 and GpGI groups, respectively, and fell into the same clusters as the homologs of D. caryophyllus, which is a member of Caryophyllaceae with G. paniculata, in phylogenetic analysis. GpFKF1 was classified into the FKF1 group in the phylogenetic tree containing the groups of FKF1 and ZTL, which has the same domain as FKF1 but a different function from FKF1. In GpFKF1, the amino acid sequences were well conserved in the three important domains of LOV, F-box, and Kelch repeats, and amino acids that are essential for the reception of blue light, interaction with GI, and ubiquitin ligase activity (Han et al., 2004; Sawa et al., 2007; Song et al., 2012) were conserved..
In A. thaliana, expression patterns of FKF1 and GI show circadian rhythms, and peaks around the dusk and in the latter half of a light period, occurring several hours before the dusk, are observed under short-day and long-day conditions, respectively (Fowler et al., 1999; Imaizumi et al., 2003). The expression patterns of GpFKF1 and GpGI in G. paniculata were similar to those of FKF1 and GI in A. thaliana. These expression patterns result in the translation and accumulation of FKF1 and GI proteins under long-day conditions, and FKF1 can interact with GI during a light period for the blue light-dependent degradation of CDF1 and its related proteins, which are the transcriptional repressors of CO and FT (Imaizumi et al., 2005; Sawa et al., 2007; Song et al., 2012). There is thus a possibility that GpFKF1 and GpGI have roles in blue light response in G. paniculata.
The LOV and F-box domains of FKF1 and the N-terminal region of GI have been reported to be necessary for their interaction (Sawa et al., 2007). The LOV and F-box domains of GpFKF1 and the N-terminal region of GpGI2 were used for the interaction assays in this study. As a result, GpFKF1 interacted with GpGI2; the interaction was also observed between the proteins from G. paniculata and A. thaliana. Because FKF1 contains the LOV domain for blue light sensing, we investigated the effect of blue light on the interaction. However, interactions were observed irrespective of the presence or absence of blue light. Further study will be needed to clarify this point. In addition to the amino acid sequences and expression patterns of GpFKF1 and GpGI, their interaction, which could be important for their function, was observed in ‘Bristol Fairy’ in the present study. Given that CRY1 and CRY2 are known to be blue light receptors in addition to FKF1 (Thomas, 2006; Wang et al., 2014), it cannot be ruled out that the variation in the blue light response of flowering is associated with the CRY-mediated flowering pathway.
Variation in the flowering response to blue light was found in G. paniculata. No defects were found in GpFKF1 and GpGI of ‘Bristol Fairy’ in this study, and its possible mechanism was associated with GpFT and GpSOC1. As one candidate, the nucleotide sequences of the promoter region of GpFT1 differed between ‘Bristol Fairy’ and ‘Million Star’, and large deletions that could affect the expression were found in ‘Bristol Fairy’. Because CO is also essential for the regulation of flowering in A. thaliana, differences in the CO homolog between the two cultivars should be investigated in G. paniculata. In contrast to the situation with red and far-red light, there have been very few useful studies of the flowering response to blue light in cut flowers. Our results will thus be useful for research and technological development in the control of flowering time in cut flowers in lighting culture and plant factories.
The authors thank Mr. T. Takahashi for his help.