2018 Volume 87 Issue 3 Pages 364-371
2018 Volume 87 Issue 3 Pages 364-371
Indonesia is one of the world’s largest fresh pepper (Capsicum spp.) fruit-producing countries, and hot peppers are essential spices in Indonesian cuisine. During the last two decades, begomovirus, which is transmitted by the whitefly, Bemisia tabaci (Gennadius), and causes pepper yellow leaf curl disease, began to cause a huge economic loss by damaging pepper plants in Indonesia. In the present study, a highly efficient inoculation method was established for Pepper yellow leaf curl Indonesia virus (PepYLCIV), the most infectious bipartite begomovirus in pepper plants cultivated in North Sumatra, by combining agroinoculation and subsequent grafting. Partial tandem repeats of PepYLCIV DNA A and B were constructed and cloned into a binary pGreenII vector, and their infectivity was tested. Co-inoculation of Nicotiana benthamiana L. and Solanum lycopersicum L. ‘Momotaro’ with PepYLCIV DNA A and DNA B resulted in the production of typical begomoviral symptoms. Both the injection of the cotyledons with cultured agrobacteria and the inoculation of the hypocotyl with agrobacterial colonies induced viral symptoms in pepper No. 218 (C. annuum L.) seedlings in approximately 55–75%. When agroinoculated symptomatic No. 218 was grafted onto an uninfected ‘Takanotsume’ (C. annuum), all newly elongated shoots from the rootstock of ‘Takanotsume’ produced typical begomoviral symptoms. Agroinoculation combined with subsequent grafting provides a highly efficient method for introducing PepYLCIV into pepper plants.
The genus Capsicum, belonging to the family Solanaceae, originated from and was first domesticated in South and Central America (Singh, 2007). Capsicum is one of the most important spices used worldwide as it accumulates pungent capsaicinoids in its fruit. It consists of several wild species and five domesticated species C. annuum L., C. chinense Jacq., C. frutescens L., C. pubescens Ruiz et Pav., and C. baccatum L. (Bosland and Votava, 2000). Of these, C. annuum, which originates from southern Mexico, is the most widely cultivated worldwide. In Southeast Asian countries, local C. frutescens cultivars are also widely grown as a crop and are present in every market. According to the Food and Agriculture Organization of the united nations statistics (FAO STAT, 2014), China, Mexico, Turkey, Indonesia, and Spain have the biggest production quantity and together produce approximately 75% of the world production of fresh pepper fruits. Hot peppers are an essential spice in Indonesian cuisine, and this country produced 1875 thousand tons of fresh pepper fruits in 2014.
According to the local farmers in North Sumatra, pepper yellow leaf curl disease (PepYLCD) caused by geminiviruses caused serious economic damage in pepper production over the last two decades. The Geminiviridae family encompasses a large number of plant viruses with circular and single-strand DNA genomes. On the basis of their genome organization and biological properties, geminiviruses are divided into the following nine genera: Begomovirus, Becurtovirus, Capulavirus, Curtovirus, Eragrovirus, Grablovirus, Mastrevirus, Topocuvirus, and Turncurtovirus. According to the International Committee of Taxonomy of Viruses (ICTV), 322 virus species are included in the genus Begomovirus, making it the largest genus of plant-infecting viruses. The majority of members of Begomovirus are bipartite, with a genome composed of two similar-sized DNA components (DNA A and DNA B) (Saunders et al., 2000). DNA A encodes a replication-associated protein (Rep), coat protein, and proteins that participate in the control of replication and gene expression. On the other hand, DNA B encodes proteins required for nuclear trafficking and cell-to-cell movement of the viral DNA. Virus infections result in a general decrease in plant growth and reduced yields, and the production is almost entirely lost if plants are infected during early growth. Whitefly, Bemisia tabaci (Gennadius), is the natural vector of begomoviruses (Cohen and Harpaz, 1964), and the B and Q biotypes of B. tabaci, in particular, have played an important role in its spread.
Over the past three decades, diseases caused by begomoviruses have contributed to production losses in solanaceous crops, particularly tomatoes (Solanum lycopersicum L.), peppers (Capsicum spp.), and eggplants (Solanum melongena L.), in many tropical and subtropical regions of the world (Kenyon et al., 2014). With the increasing international importance of leaf curl disease in tomatoes, immense efforts have been made to identify resistance mechanisms to tomato leaf curl disease and incorporate these mechanisms into improved tomato cultivars (Kenyon et al., 2014). To date, five major genes for resistance or tolerance to Tomato yellow leaf curl virus (TYLCV) (Ty-1–Ty-5) have been identified in wild tomato relatives, and some major genes have been incorporated into commercial cultivars in different areas (Anbinder et al., 2009; Hutton et al., 2012; Ji et al., 2007, 2009; Lapidot et al., 2015; Verlaan et al., 2013; Zamir et al., 1994). However, resistance to begomovirus infection is much less advanced in peppers compared to tomatoes, and there are as yet no commercial pepper cultivars carrying resistance or tolerance to begomovirus infection (Kenyon et al., 2014).
Occurrence of PepYLCD has been reported in almost all pepper growing areas in Indonesia (Trisno et al., 2009), and one of the main causal agents is known to be a bipartite begomovirus Pepper yellow leaf curl Indonesia virus (PepYLCIV) (Sakata et al., 2008; Sulandari et al., 2007; Tsai et al., 2006). Moreover, our previous study indicated that, in addition to PepYLCIV, another bipartite begomovirus, Tomato yellow leaf curl Kanchanaburi virus (TYLCKaV), and a monopartite begomovirus, Ageratum yellow vein virus (AYVV), are also associated with PepYLCD in northern Sumatra, Indonesia (Koeda et al., 2016). Thus, finding a cue to resolve this severe problem of begomovirus infection is of great importance for the optimal growth and production of horticultural crops. Besides control of whitefly, breeding for resistance against begomoviruses has been deployed as the main strategy in disease management. This approach requires appropriate and efficient screening of germplasm for begomovirus-resistant accessions. In our previous study, we established an efficient agroinoculation method for infecting tomato plants with TYLCKaV (Koeda et al., 2017). Agroinoculation can be conducted under controlled laboratory conditions using small seedlings, which can enable an accurate resistance evaluation in a short period of time and in a small space. Full-length sequences of DNA A and B of PepYLCIV, a main causal agent of PepYLCD, and successful agroinoculation of PepYLCIV to Nicotiana benthamiana L. and C. annuum have been reported (Sakata et al., 2008). However, because those studies are fundamental research on viral functions, precise information about the optimal agroinoculation method and how the pepper genotype affects the inoculation efficiency is also lacking. In the present study, a highly efficient method was established for introducing PepYLCIV into pepper plants by agroinoculation combined with subsequent grafting.
In our previous study, A6, C1, D1, and E2 pepper leaf samples were collected at Banda Aceh, Indonesia, for DNA extraction and full-length DNA A sequences of PepYLCIV were recovered (Koeda et al., 2016). In the present study, extracted DNA was used to amplify a full-length DNA B sequence using a rolling circle amplification (RCA)-based TempliPhi DNA amplification kit (GE Healthcare, Buckinghamshire, UK) according to the procedure described by Koeda et al. (2017). Nucleotide sequencing was carried out by an ABI PRISM 3100 genetic analyzer with an ABI PRISM BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, CA, USA).
Construction of infectious clonesA full-length PepYLCIV DNA A sequence (Fig. 1a), PepYLCIV-[BA_D1-1] (Koeda et al., 2016), and DNA B sequence (Fig. 1a) of PepYLCIV-[BA_D1-1-B] were cloned into pGreenII-p35S according to Koeda et al. (2017). For PepYLCIV DNA A, two fragments were amplified by PCR: fragment 1 (full-length copy of viral DNA A) with PepYLCIV fra1 F and PepYLCIV fra1 R primers, and fragment 2 (partial-length copy of viral DNA A) with PepYLCIV fra2 F and PepYLCIV fra2 R primers. Linearized pGreenII-p35S, fragment 1 and fragment 2 were ligated using an In-Fusion HD Cloning Kit (Takara, Shiga, Japan), and Escherichia coli (HST08) was transformed with a ligation product. The obtained construct was named pGreenII-p35S-PepYLCIV-DNA-A (Fig. 1b). For PepYLCIV DNA B, two fragments were amplified: fragment 1 (partial-length copy of viral DNA B) with PepYLCIV DNA-B fra1 F and PepYLCIV DNA-B fra1 R, and fragment 2 (full-length copy of viral DNA B) with PepYLCIV DNA-B fra2 F and PepYLCIV DNA-B fra2 R. Ligation and E. coli transformation were conducted in the same way as that of DNA A. The obtained construct was named pGreenII-p35S-PepYLCIV-DNA-B (Fig. 1c).

Begomovirus genomes, viral proteins, and constructed infectious clones. (a) Bipartite begomovirus has two components, DNA A and DNA B. DNA A encodes two proteins in the virion-sense AV1 (coat protein) and AV2 (pre-coat protein), and four proteins in the complementary sense AC1 (replication associated protein), AC2 (transcriptional activator protein), AC3 (replication enhancer protein), and AC4. DNA B encodes two proteins, one in the virion-sense BV1 (nuclear shuttle protein) and one in the complementary sense BC1 (movement protein). Vector maps for (b) pGreenII-p35S-PepYLCIV-DNA-A, (c) pGreenII-p35S-PepYLCIV-DNA-B, and (d) pGreenII-p35S-PepYLCIV-DNA-A+B.
To clone both DNA A and DNA B of PepYLCIV in the same pGreenII-p35S vector, the DNA B partial repeat in pGreenII-p35S-PepYLCIV-DNA-B was amplified by PCR. Amplified fragment and pGreenII-p35S-PepYLCIV-DNA-A cut by SmaΙ were ligated using an In-Fusion HD Cloning Kit (Takara), and E. coli was transformed with the ligation product. The obtained construct was named pGreenII-p35S-PepYLCIV-DNA-A+B (Fig. 1d). The primer sequences are presented in Table 1.

Primers used in this study.
Agrobacterium tumefaciens (GV2260 or EHA105) was transformed by pGreenII-p35S, pGreenII-p35S-PepYLCIV-DNA-A, pGreenII-p35S-PepYLCIV-DNA-B, or pGreenII-p35S-PepYLCIV-DNA-A+B with pSoup (Hellens et al., 2000). Frozen stock of the transformed A. tumefaciens was kept at −80°C until use.
Inoculation of Nicotiana benthamiana, tomatoes, and peppers with PepYLCIVAgroinfiltration and a colony inoculation procedure were conducted according to the method reported by Koeda et al. (2017). Agroinfiltration to N. benthamiana was conducted using pGreenII-p35S (empty), pGreenII-p35S-PepYLCIV-DNA-A, and pGreenII-p35S-PepYLCIV-DNA-B. A colony inoculation procedure to ‘Momotaro’ (Takii seed, Kyoto, Japan), a begomovirus-susceptible tomato cultivar, was conducted using pGreenII-p35S (empty), pGreenII-p35S-PepYLCIV-DNA-A, pGreenII-p35S-PepYLCIV-DNA-B, and pGreenII-p35S-PepYLCIV-DNA-A+B.
No. 218 (C. annuum) and ‘Takanotsume’ (C. annuum) (Takii seed) were utilized for PepYLCIV inoculation. In the present study, two inoculation methods were implemented. First, a colony inoculation procedure was performed using pGreenII-p35S (empty) and pGreenII-p35S-PepYLCIV-DNA-A+B. Second, agroinfiltration was conducted on the abaxial sides of the cotyledons within the two to three weeks after sowing. pGreenII-p35S (empty) and pGreenII-p35S-PepYLCIV-DNA-A+B were used for inoculation. Agrobacterium GV2260 was used for N. benthamiana and peppers, and EHA105 was used for tomatoes.
Inoculation of PepYLCIV to peppers by graftingNo. 218 was inoculated with PepYLCIV by the colony inoculation procedure. Five–six weeks after the second inoculation, the symptomatic No. 218 plants were used as scions, and PepYLCIV uninfected 8–9 weeks old seedlings of ‘Takanotsume’ (C. annuum) were utilized as rootstocks for cleft grafting. The grafted part was sealed with parafilm (Bemis Company, Inc., WI, USA), and the plants were kept in plastic bags for two weeks under humid conditions. After acclimatization, virus symptoms of lateral branches elongated from the rootstock were observed, and new developing leaves were collected for DNA extraction.
Viral DNA detection and sequence analysisNewly developed upper leaves were collected from inoculated plants, and DNA was extracted with Nucleon PhytoPure (GE Healthcare). DNA A of PepYLCIV was detected by PCR using PepYLCIV uni 2F and uni R primers. DNA B of PepYLCIV was detected using PepYLCIV DNA-B F and R primers. PCR was performed using EmeraldAmp PCR Master Mix (Takara). For the PCR reactions, the reaction mixtures were initially denatured at 94°C for 2 min, followed by 35 cycles at 94°C for 30 s, 63°C (DNA A) or 68°C (DNA B) for 30 s, and 72°C for 1 min, terminating with 3 min of extension at 72°C. The amplified PCR products were subjected to electrophoresis using 1.0% (w/v) agarose gel. The primer sequences are listed in Table 1.
Amplification of full-length viral genomes of DNA A and DNA B was conducted according to the protocol of Koeda et al. (2015) by a RCA reaction. Nucleotide sequencing was performed using an ABI PRISM 3100 genetic analyzer with an ABI PRISM BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). BLASTn (National Center for Biotechnology Information) was employed to search for the most similar begomovirus sequences in the GenBank database.
To obtain the full-length begomoviral DNA B sequence associated with yellow leaf curl symptoms, virus sequences were amplified using the RCA method. The length of the putative begomoviral DNA B sequence was 2725 nucleotides for sample A6 and C1, 2729 nucleotides for sample D1, and 2727 nucleotides for sample E2; all contained a geminiviral conserved nonanucleotide sequence (TAATATTAC) in the intergenic region (IR) and two predicted open reading frames, including one in the virion-sense (BV1) and one in the complementary sense (BC1) (Fig. 1a).
A phylogenetic tree was constructed using the sequences obtained through the BLASTn search (Fig. 2). Based on an ICTV threshold of 89% nucleotide sequence identity for demarcation of species (Fauquet et al., 2008), isolates in the present study were established as follows: The isolates BA_A6-1-B, BA_C1-1-B, BA_D1-1-B, and BA_E2-1-B were grouped with the PepYLCIV isolates (AB213599 and AB267835) from Bandung, Java, Indonesia, and sequence similarity was more than 94%. Because the full length sequences of PepYLCIV DNA A isolated from Sumatra and Java also shared high similarity (Koeda et al., 2016), it can be concluded that similar PepYLCIV isolates are widely distributed in Indonesia. The full-length sequences were deposited in GenBank (LC314792, LC314793, LC314794, and LC314795).

Phylogenetic tree based on alignments of complete begomovirus DNA B sequences. The placements of the PepYLCIV isolates in the present study are highlighted in gray. The sequences of DNA B were obtained from GenBank and used for phylogenetic analysis. The bootstrap values are indicated at the nodes (based on 1000 replicates). The branch lengths are proportional to the number of nucleotide changes as indicated by the scale bar (0.05 substitutions per site).
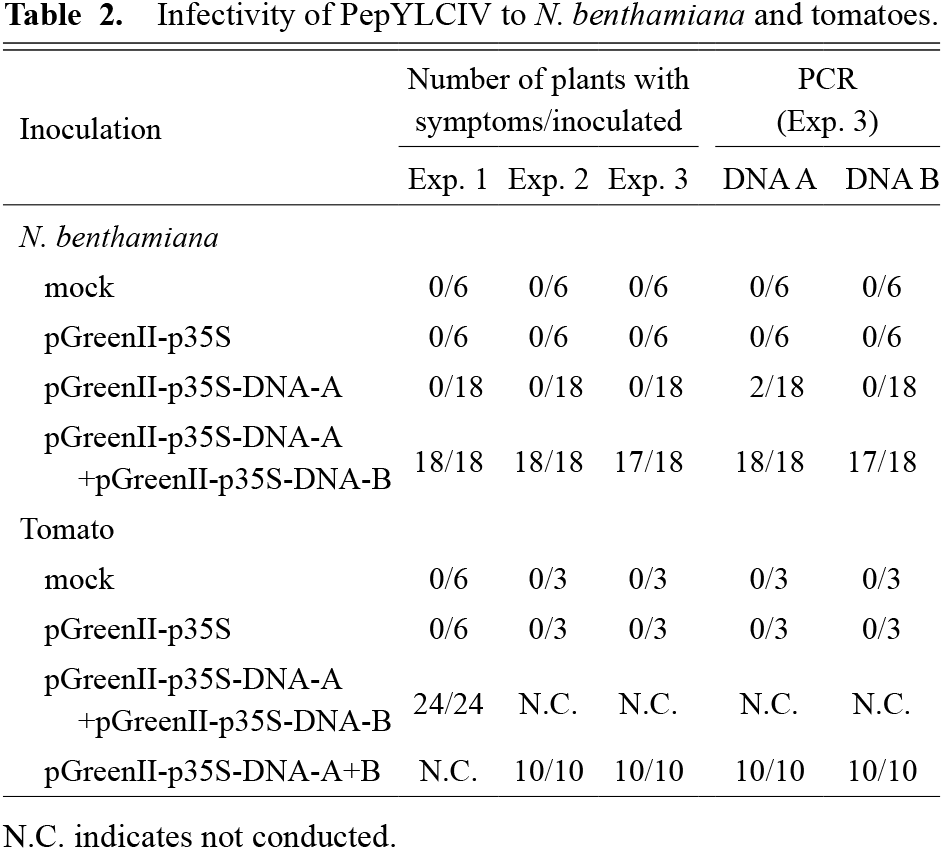
N. benthamiana and tomato plants were inoculated with the constructed infectious clones. Inoculation of buffer, empty pGreenII-p35S, or pGreenII-p35S-PepYLCIV-DNA-A alone produced no virus symptoms. Agroinfiltration of N. benthamiana with pGreenII-p35S-PepYLCIV-DNA-A and pGreenII-p35S-PepYLCIV-DNA-B produced typical begomovirus symptoms (leaf curling, yellowing, mottling, and stunting) (Fig. 3a). Similar results were obtained from independently conducted experiments. PCR was performed to examine the viral DNA A and DNA B accumulation in plants infected with infectious clones. No accumulation of either PepYLCIV DNA A or DNA B was observed in the plants inoculated with buffer or empty pGreenII-p35S (Table 2). Accumulation of DNA A was observed in several plants inoculated with pGreenII-p35S-PepYLCIV-DNA-A alone, although none of them showed typical begomoviral symptoms (Table 2). In contrast, all of the plants inoculated with pGreenII-p35S-PepYLCIV-DNA-A and pGreenII-p35S-PepYLCIV-DNA-B, accumulated PepYLCIV DNA A and DNA B (Table 2). These results showed that both components are necessary for symptom expression in N. benthamiana. Infection of N. benthamiana with bipartite begmovirus DNA A of TYLCKaV alone did not produce symptoms as also observed in our previous study (Koeda et al., 2017). Our study demonstrated that constructed clones had infectivity, and agroinoculation of PepYLCIV DNA A and DNA B can successfully induce virus symptoms.

PepYLCIV symptoms observed in N. benthamiana, tomatoes, and peppers. (a) N. benthamiana, (b) tomato ‘Momotaro’, and (c) pepper No. 218 agroinoculated with PepYLCIV. (d) Field-grown pepper plant in Indonesia infected with PepYLCIV.
Infectivity of PepYLCIV to N. benthamiana and tomatoes.
Inoculation of either buffer or empty pGreenII-p35S produced no virus symptoms in the tomato ‘Momotaro’. Conversely, agroinoculation of tomatoes with pGreenII-p35S-PepYLCIV-DNA-A and pGreenII-p35S-PepYLCIV-DNA-B or p35S-PepYLCIV-DNA-A+B produced typical begomoviral symptoms (Fig. 3b). In addition, both PepYLCIV DNA A and DNA B accumulated in all the plants with virus symptoms (Table 2). The colony inoculation procedure in tomatoes was also efficient for PepYLCIV.
Inoculation of peppers with PepYLCIVTwo methods of agroinoculation with pGreenII-p35S-PepYLCIV-DNA-A+B were applied in pepper plants. First, agrobacteria were infiltrated into the abaxial sides of the cotyledons of No. 218. To establish whether Agrobacterium cell density affects the efficiency of infection in peppers, we compared the effect of Agrobacterium cells adjusted to optical density (OD) 0.1, 0.3, 0.7, and 1.0 (Table 3). As can be seen in Table 3, the infectivity of PepYLCIV was the highest at OD of 0.1. Agroinoculation of peppers with p35S-PepYLCIV-DNA-A+B produced typical begomoviral symptoms (Fig. 3c) which resembled closely those observed in field-grown peppers in Indonesia (Fig. 3d). We further examined the inoculating stage of cotyledons with OD of 0.1. When plants were agroinfiltrated three weeks after sowing, only 7%–13% of the plants (Exp. 1, 3) showed viral symptoms (Table 4). In contrast, when plants were agroinfiltrated after two weeks after sowing, 70%–75% of plants (Exp. 2, 4) showed viral symptoms (Table 4). From these results, we can conclude that Agrobacterium cell density and inoculation stage have a crucial impact on the infectivity of PepYLCIV introduced by agroinfiltration into peppers.

Cotyledon infiltration of PepYLCIV to pepper No. 218 at various ODs.

Cotyledon infiltration of PepYLCIV to pepper No. 218 at different developmental stages.
In the present study, agrobacteria from a cultured petri dish were directly inoculated through a colony inoculation procedure. When No. 218 was inoculated, 29%–65% of the inoculated plants, with an average of 54% from 11 experiments, were infected and showed viral symptoms (Table 5). Both PepYLCIV DNA A and DNA B accumulated in all the plants with virus symptoms. Moreover, viral DNAs were recovered and sequenced from the upper uninoculated leaves. The recovered DNA A and DNA B sequences had 100% identity with the infected PepYLCIV sequence. Therefore, these results indicate that in the case of No. 218, 50%–70% of the inoculated plants could be infected with PepYLCIV by both methods, that is, agroinfiltration and the colony inoculation procedure.

A colony inoculation procedure of PepYLCIV to pepper No. 218.
Although PepYLCIV successfully infected 50%–70% of the inoculated No. 218, when ‘Takanotsume’ was inoculated using either of the two methods, only 3.5% (n = 27) of the inoculated plants were infected with PepYLCIV. The transformation efficiency of pepper cells seems to be lower than that of N. benthamiana or tomato cells, and this efficiency appears to differ depending on the genotypes of peppers.
While screening the geminivirus resistance accessions, it will be important to distinguish symptom-less resistance to inoculation-escape. For that purpose, inoculation efficiency needs to be very close to 100%. To achieve this goal, agroinoculated symptomatic No. 218 was grafted onto uninfected ‘Takanotsume’ (Fig. 4a, b). Newly elongated shoots from the rootstock ‘Takanotsume’ produced typical begomoviral symptoms (Fig. 4c, d). Moreover, the successfully grafted plants were all infected and produced viral symptoms (Table 6).

Inoculation of PepYLCIV by grafting. (a) Healthy ‘Takanotsume’ and PepYLCIV infected No. 218 from left to right. (b) PepYLCIV infected No. 218 were used as scions, and healthy ‘Takanotsume’ were utilized as rootstocks for cleft grafting. (c) Agroinoculated symptomatic No. 218 was grafted onto uninfected ‘Takanotsume’. Photograph was taken after 1 month after grafting. Lateral shoot elongated from the rootstock ‘Takanotsume’. (d) Begomoviral symptoms observed in the newly elongated lateral shoot of ‘Takanotsume’. Grafting parts are indicated with white triangles.

Infectivity of PepYLCIV to ‘Takanotsume’ by grafting.
The causal agent of PepYLCD was first identified as a begomovirus based on a partial DNA sequence, and the virus was tentatively named Pepper yellow leaf curl Indonesia virus (PepYLCIV). PepYLCIV was transmitted by a whitefly from peppers to tomatoes, ageratum (Ageratum conyzoides L.), N. benthamiana, N. glutinosa L., N. tabacum L., Vigna unguiculata L., V. radiata L., Glycine max Merr., and Physalis floridana L. (Sulandari et al., 2007). Tsai et al. (2006) determined the full-length DNA A sequence of PepYLCIV. Sakata et al. (2008) characterized the molecular structure of PepYLCIV, including its full-length DNA B sequence, and categorized it as a bipartite begomovirus. They also reported that N. benthamiana and C. annuum could be infected with the virus by agroinoculation, but the inoculation efficiency rates were approximately 50% and 38%, respectively, which are lower than that achieved by our method. In the present study, all the N. benthamiana and tomato plants, and approximately 55%–70% of the pepper plants were successfully infected with PepYLCIV by agroinoculation. Moreover, because DNA A and DNA B were cloned in the same binary vector pGreenII, all infected plants was detected in the presence of both viral components. By combining agroinoculation and subsequent grafting, all plants produced viral symptoms, aiding the screening for begomovirus-resistant germplasm. Screening for begomovirus resistance is being performed in our laboratory. In pepper-cultivating fields in Indonesia, mixed infection with PepYLCIV, TYLCKaV, and AYVV is frequently observed (Koeda et al., 2016). In our previous study, we successfully inoculated tomatoes with TYLCKaV by agroinoculation (Koeda et al., 2017). However, we did not succeed in the agroinoculation of peppers with TYLCKaV (data not shown). To further examine the synergism effect of mixed begomovirus infection, the inoculation methods need to be improved to enable the infection of a single plant with multiple begomoviruses.