2022 Volume 91 Issue 3 Pages 286-295
2022 Volume 91 Issue 3 Pages 286-295
This study was conducted to obtain insights into unreduced gamete formation in Citrus tamurana hort. ex Tanaka ‘Nishiuchi Konatsu’ and ‘Hyuganatsu’, inferred from unreduced pollen and egg cell genotype information. To establish a method for fine genotyping of tissues with higher ploidy levels (up to hexaploidy), we first identified heterozygous single-nucleotide polymorphism (SNP) markers (CiC4240-04 and CiC5327-03) with close genetic distance to centromeres of ‘Nishiuchi Konatsu’ and ‘Hyuganatsu’. Fluorescence intensity-based standard plots were generated using real-time qPCR to accurately genotype these SNP markers on a two-dimensional panel. Ten normal seeds were obtained from fertilization of unreduced gametes by self-pollination of ‘Nishiuchi Konatsu’ or ‘Hyuganatsu’ × ‘Nishiuchi Konatsu’ crosses. Inner seed coats with hexaploid endosperm tissues and tetraploid embryos were obtained from normal seeds, and their CiC4240-04 and CiC5327-03 genotypes were determined with standard plots using real-time qPCR. Genotypes of unreduced pollen and egg cells were uniquely determined from the genotype information of 6x endosperms and 4x embryos, and the unreduced gamete formation process was inferred. The results showed that unreduced pollen formation was almost entirely due to first division restitution (FDR); therefore, it was concluded that FDR was the dominant formation process for unreduced pollen in ‘Nishiuchi Konatsu’. In contrast, there was no dominant process for the formation of unreduced egg cells because both FDR and second division restitution occurred in almost the same number of individuals with unreduced egg cells.
Citrus tamurana hort. ex Tanaka ‘Hyuganatsu’ is a late citrus variety that was found as a chance seedling around 1820 in a residential area of Mr. Magata Yasutaro in Miyazaki City, Miyazaki Prefecture, Japan. Unlike other citrus species, the albedo is consumed together with the juice sacs to mitigate the sourness of the juice. Miyazaki Prefecture is a leading region for the production of this citrus, producing 53.4% (3,389 tonnes) of the total ‘Hyuganatsu’ production (6,344 tonnes) in Japan (Ministry of Agriculture, Forestry and Fisheries, 2019) followed by Kochi, Ehime, and Shizuoka prefectures. Because ‘Hyuganatsu’ exhibits strong self-incompatibility and low parthenocarpic ability, interplanting of pollinizers or hand pollination using compatible citrus cultivars are necessary for stable fruit setting and production. However, even if fertilization occurs successfully by cross-pollination, the harvested fruits contain many seeds. Because consumers do not like citrus fruits that contain seeds, some practical methods to reduce seeds in the fruits have been developed, such as gibberellic acid treatment to induce parthenocarpic fruit development without fertilization and subsequent seed formation, and pollination with pollen grains from tetraploid citrus cultivars to induce seed abortion during fruit development (Yamamoto and Iwasaki, 1994; Yamashita, 1976; Yamashita et al., 1990). However, these techniques are costly and labor-intensive. The ‘Hyuganatsu’ industry needs a new ‘Hyuganatsu’ variety that can genetically produce seedless fruits.
‘Nishiuchi Konatsu’, a bud mutation of ‘Hyuganatsu’, was found in Kochi Prefecture, Japan, in the 1980s. It was characterized as being self-compatible and exhibited seed abortion during fruit development, resulting in low-seeded fruits (Honsho et al., 2009, 2015). In our previous research, we found that ‘Nishiuchi Konatsu’ produced giant pollen grains that were approximately 1.3 times larger in diameter than those of ‘Hyuganatsu’ (Honsho et al., 2012). These giant pollen grains were considered unreduced pollen, produced from restitution because of a failed meiosis process. These unreduced pollen grains were able to overcome self-incompatibility and reach the ovule to fertilize it. After fertilization with haploid egg cells, triploid embryos were formed, but did not readily grow into normal seeds. Therefore, it is suggested that the low seed numbers observed in ‘Nishiuchi Konatsu’ were due to seed abortion after interploid hybridization. Our previous research also found that ‘Nishiuchi Konatsu’ self-pollinated fruits contained a few perfect seeds and that all seedlings of the normal seeds were tetraploid (Honsho et al., 2012). This may happen via fertilization of an unreduced egg cell, which is rarely produced in an ovule, by an unreduced pollen. After fertilization of an unreduced egg cell with unreduced pollen, a tetraploid embryo is generated and it grows successfully into a normal seed. Unreduced gametes are useful for breeding through sexual polyploidization. In citrus, several seedless triploid cultivars have been developed via sexual polyploidization using unreduced gametes (Aleza et al., 2010a, b; Cuenca et al., 2010). The genetic composition and formation mode of unreduced gametes, which are closely associated with each other, affect the breeding performance. However, there is little information about the genetic composition and formation mode of unreduced gametes in ‘Nishiuchi Konatsu’ and ‘Hyuganatsu’.
The mode of unreduced gamete formation is generally divided into two different processes: first division restitution (FDR) and second division restitution (SDR). Because these two processes result in different pairs of homologous chromosomes and (non-)sister chromatids, the mode of unreduced gamete formation is closely related to the inheritance of parental heterozygosity. Diploid gametes formed by FDR consist of non-sister chromatids, whereas those formed by SDR consist of sister chromatids (Bretagnolle and Thompson, 1995). Assuming that no crossover occurs between a certain locus and a centromere in the paired homologous chromosomes, parental heterozygosity at the locus will be completely transmitted to the 2n gametes in FDR. In contrast, if either one of the heterozygotic alleles is transmitted to the 2n gametes, it will become homozygotic in SDR (Fig. 1). In FDR, a single crossover induces 50% heterozygosity transmission and 50% homozygosity transmission of parental heterozygous alleles at a locus that is located between a centromere and a chiasma. In SDR, a chiasma located between a locus and a centromere will result in a heterozygous allele at the locus in all 2n gametes (Zhao and Speed, 1998a, b). Thus, understanding how parental heterozygosity is transmitted to unreduced gametes is necessary to estimate the mode of unreduced gamete formation.

The behavior of a pair of homologous chromosomes (2n = 2) during first division restitution (FDR) and second division restitution (SDR) in meiosis, in which a single arm exchange occurs in prophase I. In FDR, all regions between the centromere and the crossover retain parental heterozygosity, but distal regions from the crossover are either homozygous or heterozygous. In SDR, regions between the centromere and the crossover are all homozygous, and distal regions from the crossover are all heterozygous.
One method that can be used to identify genotypes of unreduced gametes is genotyping of polyploid tissues formed by the fertilization of unreduced gametes. The highest ploidy level is found in endosperm tissue as hexaploidy (2x sperm cell + two 2x polar nuclei), when unreduced female and unreduced male gametes are fertilized. Complete genotype information for 4x embryos and 6x endosperms enables determination of the genotypes of unreduced pollen and eggs (Table 1; more details are provided in the Discussion section). Therefore, methods for fine genotyping of hexaploids are required. In ‘Nishiuchi Konatsu’, tetraploid seeds can be obtained via fertilization of unreduced pollen and egg cells, either by self-pollination or by outcrossing with ‘Hyuganatsu’ × ‘Nishiuchi Konatsu’ (Honsho et al., 2012, 2015). Although the endosperm tissue generally degenerates during seed formation in citrus, it has recently been shown that the endosperm tissue remains in the inner seed coat of citrus seeds, and its ploidy level can be measured (Yanagimoto et al., 2018). This suggests that endosperm tissue DNA is available in the inner seed coat.

Possible genotypes of unreduced pollen and egg cells anticipated from genotypes of 4x embryos and 6x endosperms.
In this study, we genotyped tetraploid embryos and hexaploid endosperm tissues from seeds produced by fertilization with unreduced ‘Nishiuchi Konatsu’ pollen and ‘Nishiuchi Konatsu’ and ‘Hyuganatsu’ egg cells to identify their genotypes and infer the formation process of unreduced gametes. We first established a method to genotype cells of differing ploidy levels up to hexaploidy in citrus species. We screened for single-nucleotide polymorphisms (SNPs) in ‘Hyuganatsu’, which were located near centromeres, and where genetic recombination was suppressed. In addition, we looked for individuals that were homozygous for the obtained SNP markers from a population of ‘Hyuganatsu’ × ‘Nomabeni Hassaku’ (HY × HS) seedlings and mixed the DNA of these individuals at various ratios to create a standard sample set that simulated the possible genotypes or allele dosages, ranging from diploid to hexaploid. The allelic fluorescence intensity data obtained by SNP genotyping using a real-time quantitative polymerase chain reaction (qPCR) system were plotted on a two-dimensional panel to verify whether the mixed DNA samples could simulate allele ratios of tetraploid and hexaploid individuals. Finally, we obtained normal seeds from the fruits of ‘Nishiuchi Konatsu’ × ‘Nishiuchi Konatsu’ (NK × NK) and ‘Hyuganatsu’ × ‘Nishiuchi Konatsu’ (HY × NK) hybrids and their inner seed coat with endosperm (ISE), and embryos that germinated to seedlings were genotyped for selected SNP loci. Based on the genotype information, unreduced gamete formation of ‘Nishiuchi Konatsu’ and ‘Hyuganatsu’ was estimated.
This study examined C. tamurana hort. ex Tanaka ‘Nishiuchi Konatsu’ and ‘Hyuganatsu’, and C. hassaku hort. ex Tanaka ‘Nomabeni Hassaku’. ‘Nishiuchi Konatsu’ trees grown in a commercial orchard in Miyazaki City, and ‘Hyuganatsu’ and ‘Nomabeni Hassaku’ trees grown in the Field Science Center, University of Miyazaki, Miyazaki Prefecture, Japan were used. We also used pot-grown HY × HS seedlings.
From late April to early May in 2016, 2017, and 2018, 200 flowers each of NK × NK and HY × NK were hand pollinated. For pollen collection, flower buds just prior to anthesis were collected and undehisced anthers were detached using forceps followed by incubation at 27°C overnight to promote anther dehiscence. Before pollination, emasculation was conducted to avoid accidental self-pollination, and flowers were covered with paper bags after hand pollination to protect them from alien pollen grains. The paper bags were then removed approximately one week after pollination.
Fruits were harvested in April of the following year. Normal seeds were sampled, followed by separation of the outer and inner seed coats and embryos. The chalazal end of the inner seed coat was removed because it does not contain any ISE (Yanagimoto et al., 2018). The residual inner seed coat was further divided into two parts for ploidy measurement and DNA extraction. Naked embryos were placed on a petri dish with wet paper, and germinated seedlings were transferred to biodegradable Jiffy pots filled with vermiculite. When seedlings were grown sufficiently to allow leaf samples to be taken, a small sample of the leaf was taken for DNA extraction.
2. DNA extractionDNA extraction from ISE was performed using Illustra Nucleon Phytopure Genomic DNA Extraction Kits (GE Healthcare, Chicago, IL, USA), according to the manufacturer’s protocol. DNA extraction from leaf samples was conducted using the CTAB method (Doyle and Doyle, 1987). The obtained DNA was dissolved in 100 μL of TE buffer containing 10 μg·mL−1 RNase (Nippon Gene, Tokyo, Japan), and kept at room temperature overnight. It was then purified by phenol extraction and ethanol precipitation, followed by resuspension in 100 μL of TE buffer. The DNA concentration of all samples was measured using a Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA, USA).
3. Development of a method for fine genotyping 1) Looking for SNP markers in ‘Nishiuchi Konatsu’For SNP marker analysis, it was first necessary to look for heterozygous SNPs in ‘Nishiuchi Konatsu’ that were located near the centromere to avoid genetic recombination. Based on the available genome information for clementine mandarins listed in the Phytozome database <https://phytozome-next.jgi.doe.gov/> and a clementine genetic map (Aleza et al., 2015), 20 SNP loci located near the centromere were selected (Table 2), and sequences (including these loci) were retrieved from the Phytozome database.

Twenty SNP loci investigated in this study.
DNA was then extracted from the leaves of ‘Hyuganatsu’, ‘Nishiuchi Konatsu’, and ‘Nomabeni Hassaku’ plants using the CTAB method (Doyle and Doyle, 1987), and the selected 20 loci were amplified using PCR (T100TM Thermal Cycler; Bio Rad Laboratories, Hercules, CA, USA). PCR was performed using 1 × Emerald Amp Max PCR Master Mix (TaKaRa Bio, Shiga, Japan), 2.5 μM forward and reverse primers (Table 3), and approximately 10 ng of DNA. The reaction conditions were as follows: initial denaturation at 95°C for 1 min, followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min, and a final extension at 72°C for 2 min. Amplification was confirmed by electrophoresis on a 1% agarose gel and visualized using ethidium bromide. Amplicons were treated with ExoSAP-IT (Affymetrix, Santa Clara, CA, USA), and sequenced by direct Sanger sequencing. SNP loci that were heterozygous and found in all the cultivars studied were screened.

Primers used for PCR amplification of 20 loci for SNP identification.
Leaves were sampled from 20 HY × HS seedlings and DNAs were extracted using the CTAB method (Doyle and Doyle, 1987). The screened SNP loci were amplified using specific primer sets (Table 3) and Emerald Amp Max PCR Master Mix. After an enzymatic reaction with ExoSAP-IT, the genotype of each locus was determined by direct sequencing. Segregation was tested using a chi-square test fit of goodness, with the expected ratio being homozygote:heterozygote:homozygote = 1:2:1. Two SNP loci that fitted well to the expected ratio were selected for further experiments.
3) Making a standard plotTwo SNP loci selected from the 20 HY × HS seedlings were genotyped again using a real-time qPCR system (StepOnePlus Real-Time PCR Systems; Thermo Fisher Scientific, Tokyo, Japan). The reaction mixture consisted of 1× TaqMan® GTXpressTM Master Mix (Thermo Fisher Scientific), 900 nM primers, 200 nM probes (Table 4), and 20 ng of DNA in a total volume of 20 μL. The reaction conditions were 25°C for 30 s and 95°C for 20 s, followed by 40 cycles of 95°C for 3 s and 60°C for 20 s. Together with the results of direct sequencing, individuals with either homozygous SNP allele were selected.

Primer and probe sequences for qPCR.
For each selected individual, the DNA concentration was measured using Qubit and adjusted to 1 ng·μL−1. Two DNA solutions with different homozygous SNPs were mixed at different volume ratios of 6:0, 5:1, 3:1, 4:2, 3:3 2:4, 1:3, 1:5, and 0:6 to create a sample set that simulated different genotypes or allele dosages of tetraploid and hexaploid plants. SNP analysis was performed for each mixed DNA sample using the real-time qPCR device, and fluorescence intensities of the two SNP alleles were plotted on a two-dimensional graph. To confirm that the ratio of the two alleles in the panel was correctly plotted, ‘Hyuganatsu’ or ‘Nishiuchi Konatsu’, which are diploid and have heterozygotic SNP alleles at 1:1, were also analyzed.
Once all the data were plotted, all the plots were moved in parallel so that the coordinates of the non-template control (NTC) were located at the origin (Suppl. Fig. S1). Then, a linear transformation was performed so that the 6:0 and 0:6 samples moved to the coordinates (0, 1) and (1, 0), respectively (Suppl. Fig. S1), using them as a standard plot. In this standard plot, any point on the straight line connecting the plot of each standard sample and the origin corresponded to the allele ratio of the standard sample. Therefore, the genotype could be identified by examining the straight line closest to the plot of the sample analyzed.
4. Estimating the mode of unreduced gamete formation by fine genotyping of 4x embryos and 6x endosperms 1) FlowcytometryFor ploidy measurement, a sample was cut into 1 cm squares, and placed on a Petri dish. A chopping buffer (700 μL) was added (Yahata et al., 2010, 2015), the leaf piece was chopped 100 to 150 times with a razor, and then 55 μL of the 25 μg·L−1 propidium iodide (PI) solution was added. Ploidy levels were measured using a flow cytometer (Cell Lab Quanta SC MPL; Beckman Coulter, Brea, CA, USA).
2) Genotyping of 6x ISE and 4x embryo tissue samples to estimate the SNP genotypes of unreduced gametesThe inner seed coat of normal seeds and their germinated seedlings of NK × NK and HY × NK were investigated using real-time qPCR, together with the standard sample set for the standard plot. The fluorescence intensities of the SNP alleles were plotted on a two-dimensional panel, and genotypes were estimated by comparison with standard samples on the standard plot. Based on the genotypes of the ISE (which corresponded to endosperms) and seedlings (which corresponded to embryos), genotypes of the parental gametes (i.e., unreduced pollen and egg cells) were estimated for each sample.
PCR amplification was attempted with ‘Hyuganatsu’ and ‘Nomabeni Hassaku’ DNA for 20 loci selected from the clementine genome and genetic map information (Table 2). In the 20 loci tested, CiC0868-01 and CiC3361-04 were not successfully amplified in either ‘Hyuganatsu’ or ‘Nomabeni Hassaku’. Amplification of CiC4033-01 was not successful in ‘Nomabeni Hassaku’, and multiple fragments were observed in ‘Hyuganatsu’ and ‘Nomabeni Hassaku’ for CiC2840-01. For the other 16 loci (CiC4581-01, CiC5785-01, CiC6278-01, CiC4240-04, CiC1757-02, CiC5261-01, CiC2824-01, CiC4954-02, CiC5327-03, CiC1380-05, CiC1135-01, CiC2635-06, CiC4993-03, CiC1208-01, CiC4853-01, and CiC4620-07), only a single fragment was amplified, so direct sequencing was performed. No SNP polymorphisms between ‘Hyuganatsu’ and ‘Nomabeni Hassaku’ were found at five loci (CiC4240-04, CiC5261-01, CiC5327-03, CiC1135-01, and CiC4620-07). One or more SNP polymorphisms were detected in resting loci within a single fragment. A total of 18 SNPs were identified in CiC4240-04, CiC5261-01, CiC5327-03, CiC1135-01, and CiC4620-07, which had six, two, one, six, and three SNPs, respectively (Table 2). SNP genotypes in ‘Nishiuchi Konatsu’ can be considered the same as those in ‘Hyuganatsu’ because ‘Nishiuchi Konatsu’ is a bud sport of ‘Hyuganatsu’.
2) Selection of SNP markersSequencing was successful in 16 of the 18 SNP loci by direct sequencing of 20 HY × HS progenies. The chi-square test was used to determine whether the segregation of heterozygous SNPs at each locus fitted with the expected ratio of 1:2:1. Three SNP loci, CiC5327-03, CiC4240-04, and CiC4620-07 demonstrated good fit with the expected segregation ratio (Table 5). In this study, we decided to use the two SNP loci, CiC5327-03 and CiC4240-04, that had the best fit for subsequent analysis. Two probes were designed targeting an SNP at 226 bp (starting from the first base of the forward primer) for CiC5327-03 and two SNPs at 524 bp to 526 bp for the CiC4240-04 locus. The primer sets for these loci were designed at the flanking regions of each probe.

Segregation of SNP alleles in ‘Hyuganatsu’ × ‘Nomabeni Hassaku’ progenies.
With regard to the HY × HS seedlings, homozygotic alleles of T and C at the locus CiC5327-03 were detected in HY × HS individual 9 (hereafter expressed as HY×HS:9) and HY×HS:22, respectively. HY×HS:39 (TGG) and HY×HS:64 (CGT) were homozygous in the two linked SNPs at the 524 and 526 bp positions in CiC4240-04 (Table 5). DNA solutions adjusted to 1 ng·μL−1 were mixed at ratios of 6:0, 5:1, 3:1, 4:2, 3:3, 2:4, 1:3, 1:5, and 0:6 using two DNA solutions of HY×HS:9 and HY×HS:22 for CiC5327-03, and HY×HS:39 and HY×HS:64 for CiC4240-04. The fluorescent intensities of the two SNP alleles were projected two-dimensionally and each plot was moved and linear-transformed to make a standard plot (Suppl. Fig. S1).
2. Estimating the mode of unreduced gamete formation by fine genotyping of 4x embryos and 6x endosperms 1) Ploidy determination by flow cytometryISEs and leaves of seedlings that were germinated from embryos were obtained from 14 normal seeds found in 18 NK × NK fruits, and four normal seeds found in 13 HY × NK fruits. Ploidy level measurements revealed that nine ISEs from NK × NK normal seeds showed hexaploidy (6x) and five showed tetraploidy (4x) (data not shown). In HY × NK, three ISEs were hexaploid (6x) and one was tetraploid (4x).
The ploidy measurement of the leaves of seedlings from germinated embryos showed that eight and one NK × NK seedlings were tetraploid and diploid, respectively, and three HY × NK were tetraploid (data not shown). Because five NK × NK embryos and one HY × NK embryo did not germinate, their ploidy levels could not be measured. DNA could not be extracted from the two inner seed coats. Finally, seven and three complete sets of 6x ISE and 4x embryo DNAs were obtained from NK × NK (116, 121-1, 121-3, 136, 138, 149-1, and 169) and HY × NK (61, 69, and 83), respectively, for further SNP genotyping by qPCR.
2) GenotypingGenotyping analysis was performed for the samples confirmed to have 6x endosperms and 4x embryo tissues using flow cytometry. Based on real-time qPCR analysis of these samples and the standard samples, the plots of samples were placed near either one of the lines connecting the origin and the plot of standard samples on the standard plot to determine the SNP genotype of each sample (Fig. 2). For example, the genotypes of CiC5327-03 for the NK×NK:116 ISE and embryo were determined to be TCCCCC and TCCC, respectively. The genotyping results for a total of nine individuals with 6x endosperms and 4x embryos are shown in Table 6.

Determination of genotypes of tetraploid and hexaploid tissues using standard plots of CiC5327-03 (top) and CiC4240-04 (bottom) SNP markers. Cross marks indicate standard samples reflecting each allelic configuration. Triangles indicate diploid leaves of ‘Nishiuchi Konatsu’. Filled circle symbols indicate samples tested.
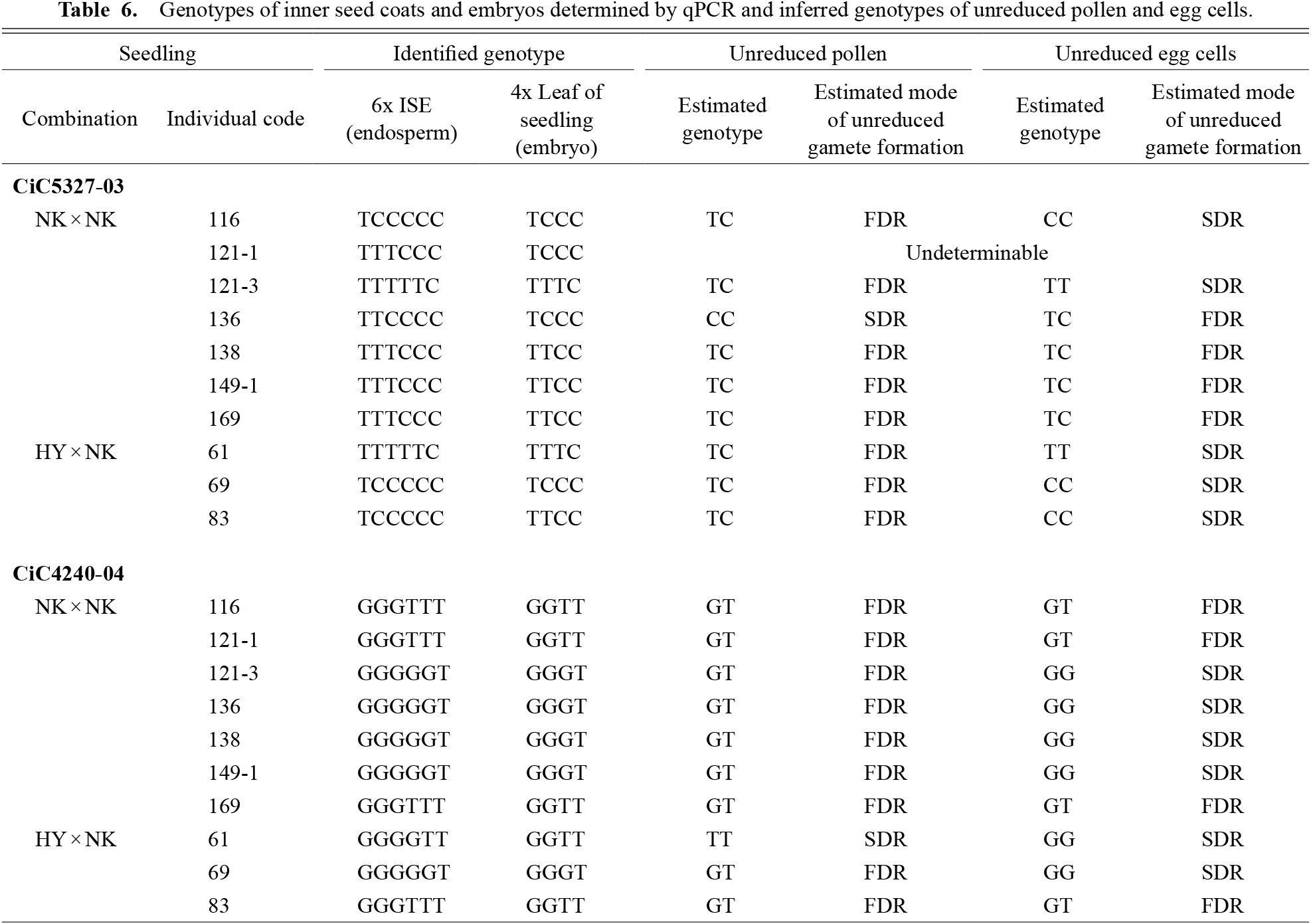
Genotypes of inner seed coats and embryos determined by qPCR and inferred genotypes of unreduced pollen and egg cells.
There was a close relationship between the formation mode and genotype at the locus near the centromere of unreduced gametes (Fig. 1). To determine the genotype of unreduced gametes, the genotype of 4x embryos (germinated seedlings) and 6x endosperm tissues, which were formed through fertilization between unreduced pollen and egg cells, were determined. If genotypes of both 4x embryos and 6x endosperms can be identified, genotypes of unreduced pollen and egg cells can also be determined. For example, assuming that the genotype of the obtained 4x embryos is aaab at a certain locus, the possible genotypes of the unreduced pollen and egg cells are aa or ab; the genotypes of the unreduced gametes cannot be determined from the embryo genotype alone. If the genotype of the 6x endosperms is additionally determined as aaaaab, then the genotype of the unreduced pollen and egg can be uniquely determined as ab and aa, respectively (Table 1). As the genotype of unreduced gametes is associated with the process of unreduced gamete formation (Fig. 1), genotypes of 4x embryos and 6x endosperms can provide information about the process of unreduced gamete formation.
In this study, we were able to identify genotypes of the 6x endosperms from ISE tissue and the 4x embryos from leaves of the germinated seedlings of nine individuals. Genotypes of unreduced pollen and egg cells were successfully estimated in all individuals, except NK×NK:121-1 (Table 6). In NK×NK:121-1, the genotype at locus CiC5327-03 was TTTCCC and TCCC for the endosperm and embryo, respectively. Since there was no possible genotype combination of unreduced pollen and egg cells that corresponded with the result of the endosperm and embryo genotypes (Table 1), its genotypes could not be estimated from this locus (Table 6).
The results of this study showed that almost all of the unreduced pollen were heterozygous (Table 6). Since both loci analyzed in this study were expected to be located close to the centromere (Aleza et al., 2015), transmission of parental heterozygosity to unreduced pollen suggested that unreduced pollen formation was due to the FDR process (Fig. 1). The critical role of the FDR process in unreduced pollen formation in this study corresponded with the observations of our previous study (Honsho et al., 2016). However, only CiC5327-03 of NK×NK:136 and CiC4240-04 of HY×NK:61 were estimated to be homozygous for unreduced pollen. Because CiC5327-03 and CiC4240-04 are located at a genetic distance of 8.2 cM and 9 cM from the centromere, respectively (Aleza et al., 2015), it is possible that the genetic recombination at CiC5327-03 and CiC4240-04 in NK×NK:136 and HY×NK:61, respectively, resulted in homozygotic alleles. Likewise, the estimated modes of unreduced gamete formation from the two SNP loci did not concord in the six cases (Table 6), which may be due to genetic recombination. The genetic distance of each marker from the centromere in ‘Hyuganatsu’ may be much larger than that reported in clementine.
In addition to unreduced pollen, genotypes of unreduced eggs were also estimated in this study: in CiC5327-03, four individuals were heterozygous and five were homozygous for unreduced egg cells, and in CiC4240-04, four individuals were heterozygous and six were homozygous, suggesting that unreduced egg cells were formed by both FDR and SDR processes (Table 6). In contrast to the FDR-dominant production of unreduced pollen shown in this study and the one exclusively found in ‘Nishiuchi Konatsu’ (a bud sport of ‘Hyuganatsu’) by Honsho et al. (2016), unreduced egg formation in ‘Hyuganatsu’ and ‘Nishiuchi Konatsu’ occurred by chance, resulting in an almost 50% probability of FDR and SDR occurrence. Thus, it was suggested that unreduced egg formation was not genetically controlled in ‘Hyuganatsu’ and ‘Nishiuchi Konatsu’.
These 2n gametes are considered useful for polyploid breeding. The advantages of FDR and SDR gametes in polyploid breeding have been discussed previously: unreduced gametes produced by FDR that have pairs of non-sister chromatids transmit more parental heterozygosity to offspring than unreduced gametes produced by SDR (Geng et al., 2019; Han et al., 2018; Liesebach et al., 2015). Therefore, FDR gametes are effective in producing offspring that maintain parental heterozygosity and epistatic interactions (Mendiburu and Peloquin, 1977a, b). However, SDR can produce polyploid offspring with more diverse multi-locus genotypic combinations and can produce populations with many polymorphisms (Aleza et al., 2016; Cuenca et al., 2011). For 2n female gamete formation in citrus, SDR is dominant in mandarin (Cuenca et al., 2015) and lemon (Rouiss et al., 2017b); while Ferrante et al. (2010) indicated that different formation modes were found depending on the cultivar. The formation of unreduced 2n pollen in citrus has been reported to be dominated by FDR for ‘Nishiuchi Konatsu’, as reported in this study and in our previous study (Honsho et al., 2016), or by both FDR and SDR for a diploid hybrid tangor (Rouiss et al., 2017a). Because ‘Nishiuchi Konatsu’ has a characteristic high-frequency production (Honsho et al., 2012) of FDR-dominant unreduced pollen, it will be useful for polyploid breeding to produce seedless triploid citrus cultivars.
The authors would like to thank Mr. Makoto Nakamura and staff at the Field Science Center at the University of Miyazaki for providing plant materials.