2022 Volume 91 Issue 3 Pages 375-381
2022 Volume 91 Issue 3 Pages 375-381
Anthracnose, caused by the fungal pathogen Colletotrichum orbiculare, is one of the most severe diseases in watermelon (Citrullus lanatus) cultivation in Japan. Genetically conferred host resistance is the best way to control it. To develop new cultivars with high resistance, identification of promising sources of resistance is important. We focused on an old Japanese cultivar, ‘Tanso Teikosei’ (NR28), with high resistance, preserved at the Nara Prefecture Agricultural Research and Development Center, evaluated its resistance to diverse C. orbiculare strains distributed in Japan, and revealed the inheritance of its resistance. NR28 and two other accessions were resistant to the highly virulent MAFF 306737 strain. NR28 was also resistant to 16 other strains collected from various regions of Japan. Many strains caused severe symptoms and wilting in susceptible accessions, including the popular Japanese F1 cultivar WF01. CAPS marker analysis indicated that NR28 and the two other resistant accessions were homozygous for the resistance allele of Cla001017, previously reported as the gene responsible for resistance to US race 1. Susceptible accessions were homozygous for the susceptibility allele at Cla001017. Segregation analyses using F2 and BC1F1 lines derived from crosses between NR28 and a susceptible accession suggested that the dominant mutant allele of Cla001017 caused the high resistance of NR28. In contrast, NR28 and other resistant accessions homozygous for the resistance allele of Cla001017 were highly susceptible to two strains sampled in Iwate Prefecture. This suggests that these strains have overcome the Cla001017 resistance allele, as have strains of C. orbiculare classified as US race 2. NR28 and the two other resistant accessions are promising materials for breeding resistance in diverse C. orbiculare strains distributed in Japan that have similar pathogenicity to US race 1. The CAPS marker that we designed in this study offers efficient analysis for watermelon breeding.
Watermelon (Citrullus lanatus (Thunb.) Matsum. et Nakai) is one of the most important cucurbits worldwide. It is generally grown in open fields from early spring to summer. Open-field cultivation is cost effective, but plants are more prone to disease and pests than those in greenhouses, where it is relatively easy to control conditions. Anthracnose, caused by the fungal pathogen Colletotrichum orbiculare (Berk. et Mont.) Arx (syn. C. lagenarium (Pass.) Ellis et Halst.), is one of the most common diseases in watermelon open-field cultivation and is the most critical target to control in watermelon production in Japan. This fungus infects watermelon and many other cucurbit crops in warm and humid regions of the world, including Japan. Anthracnose development is promoted at temperatures between 20 and 27°C under high humidity. Spread of C. orbiculare spores and infection depend upon water droplets such as rain and irrigation spray. Seed transmission is a component of its cycle. The fungus can infect aboveground organs, such as leaves, stems, and fruits. Infected areas on leaves and stems turns into brown or black lesions. In cases of severe infection, many lesions become larger, and the plants die. Black circular or elongated lesions occur on fruit, producing a gelatinous mass of pink or orange spots (Wehner et al., 2020). Thus, it affects not only fruit yield, but also fruit quality. To control the disease, sterilization of seeds, application of fungicides, and low-temperature storage of fruits after harvest are used. However, it is difficult to completely control the disease using these methods, and introduction of resistant cultivars is vital.
The USA is ahead of Japan in the development and use of anthracnose-resistant cultivars. In the USA, seven races of cucurbit anthracnose have been reported, of which races 1 and 2 appear to be the most common (Goode, 1956; Jenkins et al., 1964; Sitterly, 1973; Wasilwa et al., 1993). US race 1 is highly virulent in cucumber, cantaloupe melon, and some watermelon cultivars, whereas US race 2 is moderately virulent in most cucumber cultivars and highly virulent in most watermelon cultivars (Wasilwa et al., 1993). Since the mid 19th century, many watermelon cultivars have been developed in the USA (Levi et al., 2001). Among them, two leading cultivars, ‘Charleston Gray’ and ‘Crimson Sweet’, which are resistant to US races 1 and 3, have been used in numerous watermelon breeding programs (Suvanprakorn and Norton, 1980; Jang et al., 2019). ‘AU-Producer’, an inbred line derived from a cross between ‘Crimson Sweet’ and ‘PI 189225’, was released as being resistant to US race 2 (Norton et al., 1985). Many accessions with resistance to anthracnose have been identified; their resistance genes are named Ar-1 (against US races 1 and 3) and Ar-2 (against US race 2) (Guner and Wehner, 2003). Recently, Jang et al. (2019) reported that a non-synonymous SNP in Cla001017, which encodes a coiled-coil nucleotide-binding site leucine-rich repeat (CC-NBS-LRR) protein, results in an arginine-to-lysine substitution in the LRR domain and confers resistance to US race 1. This mutation is conserved among various cultivars resistant to US race 1, including ‘Charleston Gray’ and ‘Crimson Sweet’.
In Japan, almost all cucurbits, except squash, have been damaged by C. orbiculare strains. In particular, the production of cucumber and watermelon has been severely affected by anthracnose for a long time. However, the development and use of resistant cultivars of both crops have not progressed much: no cucumber cultivar has high resistance, and only one watermelon cultivar has been commercialized. Recently, Sano et al. (2019) evaluated anthracnose resistance in watermelon genetic resources preserved at the Nara Prefecture Agricultural Research and Development Center, and reported 16 accessions with moderate to high resistance to a Japanese anthracnose strain. Although race differentiation of C. orbiculare has not been confirmed, there is wide variation in virulence among strains in Japan (Matsuo et al., 2022). Therefore, to accelerate breeding using these accessions and to efficiently develop new cultivars with high anthracnose resistance, we must re-evaluate resistance to a wide range of C. orbiculare strains, clarify the inheritance of this resistance, and establish a marker-assisted selection (MAS) technique.
Here, we focused on an old Japanese cultivar, ‘Tanso Teikosei’, with the highest resistance among the 16 accessions reported by Sano et al. (2019), and re-evaluated its resistance to the C. orbiculare strain MAFF 306737, which causes severe damage to cucumber, and to 18 other C. orbiculare strains collected from various regions of Japan. To clarify the inheritance of the resistance, we evaluated the resistance of progeny derived from a cross between resistant and susceptible accessions. In addition, we designed a CAPS marker for the previously reported resistance mutation of Cla001017 and genotyped the accessions and progeny to clarify the relationship between this gene and the phenotypes of inoculated plants.
We re-evaluated the anthracnose resistance of a resistant old Japanese cultivar, ‘Tanso Teikosei’ (NR28); two other resistant accessions, ‘Colorad Preserving Citron’ (NR114) and ‘Wiet Resistant Congo’ (NR149); three susceptible accessions, ‘Asahi Yamato’ (NR3), ‘Shin Yamato 1’ (NR23), and ‘Turkmenskij Mramornyj’ (NR145), preserved at the Nara Prefecture Agricultural Research and Development Center; and a popular Japanese F1 cultivar (WF01). To investigate the inheritance of resistance, we generated F1s by reciprocal crossing between NR28 and NR145 [F1A, NR28 (♀) × NR145 (♂); F1B, NR145 (♀) × NR28 (♂)]. We obtained F2 lines by selfing of F1A, and BC1F1 by backcrossing of F1A (♀) × NR145 (♂).
Fungal materials and preparation of inoculumWe used 15 strains of C. orbiculare collected from all over Japan and preserved in the Genebank of the National Agriculture and Food Research Organization (NARO) (Table 1). We also used four strains (IT21-1, IT21-2, ST21-1, and ST21-2) isolated from watermelon and cucumber leaves (Table 1). Strains were cultured on potato dextrose agar (PDA; Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) at 25°C under light. Strains that did not form conidia by this method were cultured on PDA under UV light at 25°C, or on oatmeal agar (Becton, Dickinson and Company, NJ, USA) at 25°C in the dark. Mycelial plugs with conidia were soaked overnight in 10% glycerol (Fujifilm Wako Pure Chemical Corporation, Osaka, Japan) and then stored at −80°C. For inoculum preparation, a mycelial plug was transferred onto PDA and incubated at 25°C under light for 10 days. The mycelium and spores were then suspended in deionized water and spread on other PDA plates. These plates were then incubated at 25°C under light for 5–7 days. Spores formed on the surface were suspended in deionized water and filtered through Kimwipe tissue (Nippon Paper Crecia Co., Ltd., Tokyo, Japan). The suspension was shaken well. Spores were counted with a hemocytometer, their concentration was adjusted to 1.0 × 106 spores·mL−1, and 0.5 μL·mL−1 Tween-20 (Kanto Chemical Co., Inc., Tokyo, Japan) was added.
List of C. orbiculare strains used in this study.
The resistance of all accessions and cultivars was evaluated by a seedling assay with the highly virulent strain MAFF 306737. Twenty seedlings per accession/cultivar were tested in two experimental replications. To make all accessions germinate at the same time, the seeds were soaked overnight in 30°C water in a shaker. The seed coat of accessions with a thick seed coat was scratched with a nail clipper before soaking. Then, seeds were placed in Petri dishes lined with wet filter paper (Toyo Roshi Kaisha, Ltd., Tokyo, Japan) and incubated at 28°C in the dark for a day. Germinated seeds were transplanted into 6-cm-diameter plastic pots filled with moist culture soil (Nihon Hiryo Co., Ltd., Fujioka, Japan) and incubated at 28°C under a 12-h photoperiod in a climate chamber (Nippon Medical and Chemical Instruments Co., Ltd., Osaka, Japan). After cotyledon unfolding, the chamber’s temperature was set to 25°C during the day (12 h) and 18°C at night for 18 days. Plants were put inside of clear plastic boxes (16 per box; Akasaka Co., Ltd., Atsugi, Japan), and inoculum was sprayed on the upper surface of each true leaf (ca. 0.4 mL) from a spray bottle. The boxes were then closed to maintain 100% relative humidity and kept in the dark for 24 h. The lids were then opened, and the plants were grown at 25°C under a 12-h photoperiod in the climate chamber. The resistance of each inoculated plant was scored one week after inoculation. Scoring was based on infection severity (IS) levels: 0, no lesions; 1, a few small lesions (< 3 mm in diameter); 2, several small lesions or a few medium lesions (3–5 mm); 3, many small lesions and a few to several medium lesions, or a few large lesions (> 5 mm) with chlorotic boundaries; 4, several medium lesions and a few large lesions with chlorotic boundaries; 5, many large lesions over half or more of the true leaf; 6, true leaf completely withered; 7, whole seedling wilted (dead).
The resistance of NR28 and WF01 was further evaluated by seedling assays with the other 18 strains. In each trial, 20 seedlings per accession/cultivar were tested in two experimental replications as above, but seedlings were grown in plastic plug trays (32-mm-diameter holes). In addition, the resistance of NR28, NR145, and their progeny (reciprocal F1s, F2, and BC1F1) was tested by seedling assays with MAFF 306737. The resistance of each plant was evaluated as either susceptible (IS = 7) or resistant (surviving).
Fruit assayA fruit assay was performed once to evaluate symptoms on the fruit surface of NR28, NR145, and their F1A. Seeds were sown in 6-cm-diameter plastic pots filled with moist culture soil on June 10th, 2021, and seedlings were grown for 14–20 days in a greenhouse. The seedlings were then transplanted into rectangular planter boxes (60 L) filled with Peat Mix (Hokkaido Peatmoss Inc., Konosu, Japan) and slow-release fertilizer (N:P:K = 13:9:11; JCAM Agri. Co., Ltd., Tokyo, Japan), and placed in an open field. Flowers were pollinated artificially in the morning and fruits were harvested 25 days after pollination later in August. Five fruits per accession/F1A were then incubated at room temperature for five days. Fruits were put on Petri dishes inside of clear plastic boxes, 3 per clear plastic box, and the boxes were moistened with a little water to keep the humidity high. Then MAFF 306737 inoculum (1.5 × 106 spores·mL−1) was sprayed on the upper surface of each fruit (ca. 2.0 mL) from a spray bottle. The boxes were closed to maintain 100% relative humidity and kept in the dark for 48 h. The lids were then opened, and the fruits were kept at 25°C under a 12-h photoperiod in a climate chamber. The symptoms of each inoculated fruit were evaluated one week after inoculation.
Detection of polymorphism at Cla001017 by a CAPS markerGenomic DNA of each plant of NR28 and NR145 and their progeny was extracted from cotyledons using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocols with minor modifications. To determine whether the Cla001017 genotype of each plant was wild type (susceptible) or mutant (resistant), we used a cleaved amplified polymorphic sequence (CAPS) marker designed with a 300-bp amplicon size by Primer3plus <https://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi> and the Oligo calculator <http://www.ngrl.co.jp/tools/0217oligocalc.htm>: forward primer 5'-GCTTCCAAAATCTATTGTTTTGCT-3'; reverse primer 5'-TTTTTACTTCCAACACGCTCG-3'. PCR mixtures (5 μL) contained template DNA of each sample, Blend Taq-Plus, 10 × buffer, 2 mM dNTPs (Toyobo Co., Ltd., Osaka, Japan), autoclaved Milli-Q water, and primer mix. The PCR conditions were 94°C for 2 min; 35 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 30 s; and a final extension at 72°C for 3 min. Amplified DNA was digested with HindIII (Takara Bio Inc., Kusatsu, Japan) in 10 × M buffer and autoclaved Milli-Q water at 37°C for 2 h. The digested products were electrophoresed in 8% polyacrylamide gel. The gel was stained with 0.03 μL·mL−1 GelRed (Cosmo Bio Co., Ltd., Tokyo, Japan) and visualized under UV light. The Cla001017 genotype of each plant was determined from the band size: the digested band (250 bp) was the mutant allele, and the undigested one (300 bp) was the wild-type allele.
The three resistant accessions (NR28, NR114, NR149) survived when inoculated with the highly virulent strain MAFF 306737, and their average IS scores ranged between 2.6 to 4.2; the other 3 accessions and WF01 wilted and died, and their average IS scores were 7.0 (Fig. 1). NR28 had a significantly lower IS score than other accessions, and thus the highest resistance (Fig. 1A). NR28 had lower IS scores than WF01 in trials using most strains except for IT21-1 and IT21-2, and significantly so in 9 strains (MAFF 306795, 726522, 306519, 306738, 237606, 306870, and 306737, ST21-2, and ST21-1) (Fig. 2A). These nine strains caused all or some WF01 seedlings to wilt, but NR28 seedlings survived with only a few small lesions (Fig. 2B). Strains IT21-1 and IT21-2 caused severe necrotic symptoms on the leaves of both NR28 and WF01, and many plants wilted (IS = 7.0) (Fig. 2A). The remaining eight strains (MAFF 240422, 306590, 306868, 306518, 306685, 306681, 243071, and 242678) caused no or minor symptoms on the leaves of both NR28 and WF01. In the fruit assay, the fruit surface of NR28 had no or minor lesions, but that of NR145 had many large lesions with salmon-colored spores (Fig. 3A). The F1A fruits had an intermediate phenotype, with a few large lesions and many small lesions. The symptoms appeared only on the skin, with no browning or decomposed flesh.

(A) Distributions of infection severity scores of six accessions and one commercial F1 cultivar (WF01) in true leaves. Boxplots with the same letter are not significantly different at P < 0.05 by Tukey’s HSD; vertical bars indicate SD. (B) Seedlings of four accessions inoculated with C. orbiculare strain MAFF 306737. (C) fragment patterns of the Cla001017 CAPS marker.
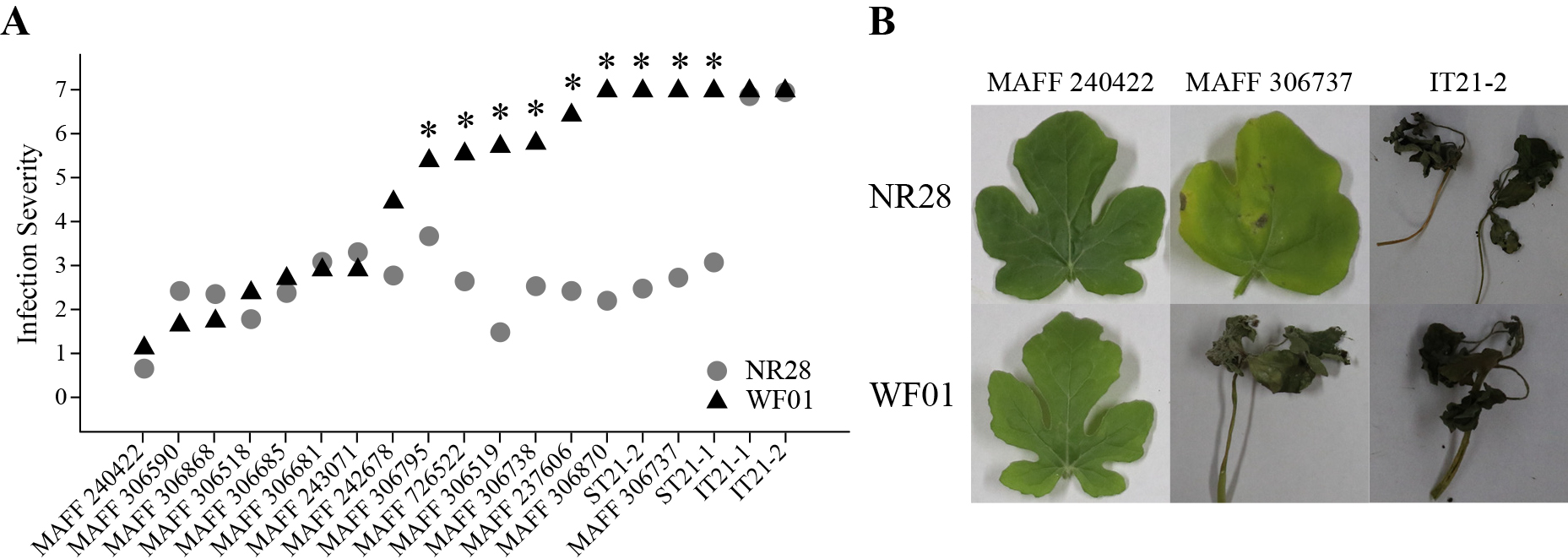
(A) Distributions of infection severity scores of WF01 and NR28 in true leaves and seedlings inoculated with 19 C. orbiculare strains. *Significant difference between WF01 and NR28 at P < 0.05 by Tukey’s HSD. (B) True leaves or seedlings inoculated with C. orbiculare MAFF 240422, MAFF 306737, and IT21-2.

Fruits and seedlings of NR28, NR145, and their F1A inoculated with MAFF 306737.
CAPS marker analysis indicated that the three resistant accessions (NR28, NR114, NR149) were homozygous for the resistance (mutant, “mut”) allele of Cla001017 (Fig. 1C). In contrast, the susceptible accessions (NR3, NR23, NR145) and WF01 were homozygous for the susceptibility (wild-type, “WT”) allele (Fig. 1C). Reciprocal F1s between NR28 and NR145 were thus heterozygous (mut/WT), and all seedlings were assigned to the resistant phenotype (Table 2). Among 104 BC1F1 seedlings derived from F1A × NR145, 56 were heterozygous (mut/WT), of which 55 were assigned to the resistant phenotype (Table 2). In contrast, 48 seedlings were homozygous (WT/WT), and all were assigned to the susceptible phenotype. The segregation ratios of both genotype (χ2 = 0.615, P = 0.433) and phenotype (χ2 = 0.346, P = 0.556) fitted the expected value (1:1). In the NR28 × NR145 F2 population, 21 mutant-type homozygous seedlings (mut/mut) and 47 heterozygous seedlings (mut/WT) were assigned to the resistant phenotype. Two heterozygous (mut/WT) and 29 wild-type homozygous seedlings (WT/WT) were assigned to the susceptible phenotype. The segregation ratios of both genotype (χ2 = 1.303, P = 0.521) and phenotype (χ2 = 2.104, P = 0.150) fitted the expected values (1:2:1 and 3:1, respectively). In addition, there were significant associations between genotype and phenotype in both the BC1F1 and F2 populations (P < 0.001; Fisher’s exact test).

Segregation of resistance to anthracnose strain MAFF 306737 in NR28, NR145, their reciprocal F1s (F1A, NR28 ♀ × NR145 ♂; F1B, NR145 ♀ × NR28 ♂), BC1F1 (F1A ♀ × NR145 ♂), and F2 (selfing of F1A).
We re-confirmed that NR28, a candidate source of anthracnose resistance for breeding, has strong resistance to most strains present in Japan. In contrast, many strains caused severe symptoms and wilting of susceptible accessions, including the popular Japanese F1 cultivar WF01. Genotyping of Cla001017 showed that all resistant accessions were mut/mut, and susceptible accessions were WT/WT (Fig. 1C). This suggests that the high resistance was caused by the mutant allele of Cla001017. In the progeny of the NR28 × NR145 cross, the phenotype fitted a 3:1 ratio in the F2 generation and a 1:1 ratio in the BC1F1 generation, consistent with the genotype of Cla001017. These results strongly suggest that the high resistance of NR28 and other resistant accessions was due to Cla001017. The equivalent resistance at the seedling stage between plants of mut/mut and mut/WT in the F2 generation suggests that the resistance of the mutant allele was dominantly inherited. This is consistent with the report by Jang et al. (2019) that the dominant Cla001017 mutant allele contributed the resistance to C. orbiculare US race 1 (Wasilwa et al., 1993).
Jang et al. (2019) identified Cla001017, annotated as encoding a CC-NBS-LRR (NBS-LRR) resistance protein. Most NBS-LRR genes are expressed during infection and possibly contribute to the recognition of effectors, and can in turn enhance resistance through immune responses (Kourelis and van der Hoorn, 2018). Therefore, Cla001017 is probably responsible for resistance to C. orbiculare. Interestingly, the resistance of NR28 × NR145 F1 seedlings heterozygous (mut/WT) for Cla001017 was as high as that of homozygous (mut/mut) seedlings (Fig. 3B), but fruits developed large lesions on the skin (Fig. 3A). This difference indicates that the Cla001017 mutant allele works dominantly at the seedling stage, but its effect in fruit decreases after harvest. The expression level of Cla001017 may differ between fruit and other organs (hypocotyls and leaves), possibly leading to differences in lesion formation between them. Any new F1 cultivar should be homozygous for the Cla001017 mutant allele, and thus breeding to introduce the mutant allele into both parental lines is important.
The resistant accession NR28 had low IS scores and stable resistance to all but two C. orbiculare strains (Fig. 2A). There were no significant differences between NR28 and WF01 when inoculated with eight strains (MAFF 240422, 306590, 306868, 306518, 306685, 306681, 243071, and 242678), but WF01 was significantly less resistant than NR28 when inoculated with nine strains (MAFF 306795, 726522, 306519, 306738, 237606, 306870, and 306737, ST21-2, and ST21-1) (Fig. 2A). This result suggests that the former strains were incompatible with both NR28 and WF01, while the latter were compatible with WF01 but not NR28, and thus race differentiation occurs in C. orbiculare in Japan. Thus, the former strains might be regarded as a race that does not adapt to watermelon, although the previous study did not mention this. As the mutant allele of Cla001017, known to be incompatible with US race 1, confers the high resistance of NR28, this suggests that the latter nine strains correspond to US race 1. In contrast, NR28, homozygous for the Cla001017 mutant allele, had high IS scores when inoculated with IT21-1 and IT21-2. This suggests the occurrence of a race in Japan similar to US race 2 that can overcome the effect of the Cla001017 mutant allele. However, since this study used only two watermelon accessions, more evidence is required to correctly differentiate C. orbiculare races in Japan. In a future study, it will be necessary to examine the compatibility of the strains with the Cla001017 mutant allele using many other watermelon cultivars to differentiate the C. orbiculare races in Japan. As resistant cultivars harboring the Cla001017 mutant allele become widespread in Japan, strains that overcome this allele such as IT21-1 and IT21-2 may come to dominate. Thus, in preparation for such a situation, it is vital to discover new genetic resources with high resistance to such strains.
NR28 and other resistant accessions harboring the Cla001017 mutant allele are promising materials for breeding cultivars with resistance to diverse C. orbiculare strains with pathogenicity similar to that of US race 1. However, fruit quality and other important agronomic traits differ between almost all genetic resources and current Japanese commercial F1 cultivars. This indicates that a lot of effort will be needed to develop new cultivars with both high anthracnose resistance and high quality fruit. Fortunately, like the CAPS marker designed in this study, the sequence information on the Cla001017 mutant allele can be used to select resistant individuals or lines from numerous materials in breeding programs. Therefore, the development of new resistant cultivars can be promoted efficiently, and introducing these cultivars into production areas in Japan is expected to reduce the damage caused by anthracnose.
We thank Y. Ishiga, N. Nishi, and Z. Shuang of the University of Tsukuba and Y. Kubo of Setsunan University for technical advice and assistance. We are grateful to A. Mochizuki and M. Amano of Saitama Gensyu Ikuseikai Co., Ltd., and J. Shimazu and R. Tsujita of Nanto Seed Co., Ltd., for providing infected plant materials from producing areas in Japan. Most fungal strains were kindly provided by the NARO Genebank.