2024 Volume 93 Issue 3 Pages 242-250
2024 Volume 93 Issue 3 Pages 242-250
Columnar-shaped apple (Malus × domestica Borkh.) trees are of interest because of their profitable and labour-saving characteristics based on high-density planting and robotic technologies. Recent studies have shown that the Co gene, which is responsible for the columnar shape, and MdPG1, which contributes to fruit storability and flesh texture, are located on chromosome 10 of the apple genome. In silico analysis and genotypic characterisation of our columnar breeding materials revealed that the unfavourable haplotype carrying both Co and the MdPG1-3 allele that confers low storability was retained in almost all of our current and historical columnar materials because of the strong linkage between the two genes. To overcome this limitation, we conducted large-scale marker-assisted selection to obtain plants harbouring a favourable haplotype conferring a columnar tree with improved storability, and high-quality flesh resulting from recombination events between these genes during meiosis. Large-scale marker-assisted selection of approximately 15,000 seedlings composed of five crosses resulted in the identification of 80 individuals harbouring the targeted recombinant haplotype. These individuals are valuable for breeding columnar apple cultivars with superior fruit quality.
Columnar tree shape of apples (Malus × domestica Borkh.) differs from the normal tree shape in terms of shoot morphology, including short internodes and reduced branching. It was discovered in Canada in the 1960s as a spontaneous mutant of ‘McIntosh’ (Fisher, 1995). It has gained interest because of its profitable and labour-saving characteristics that leverage high-density planting and robotic technologies (Moriya-Tanaka et al., 2023). The National Agriculture and Food Research Organization (NARO) has initiated columnar apple breeding through a crossbreeding strategy since the 1980s, as it is dominant in inheritance over normal tree shapes owing to the expression of the Co gene (Lapins, 1976).
The molecular mechanisms underlying the columnar tree shape were also investigated. Genetic mapping narrowed the candidate region to a few hundred kilobases (kb) on chromosome 10 (Chr10) (Bai et al., 2012; Baldi et al., 2013; Moriya et al., 2012). A nucleotide sequence comparison of the causal region using bacterial artificial chromosome libraries of ‘McIntosh’ and a columnar cultivar ‘Telamon’ identified an 8-kb retrotransposon insertion, which is the causal mutation for the columnar trait (Okada et al., 2016; Otto et al., 2014). Analysis of differentially expressed genes between columnar and normal trees identified a candidate gene, MdDOX-Co, which encodes a 2-oxoglutarate-dependent dioxygenase located 16-kb downstream of the insertion mutation (Okada et al., 2016; Petersen et al., 2015; Wolters et al., 2013). The short internode lengths observed from MdDOX-Co overexpressed transformants using Arabidopsis, tobacco, and apple confirmed that MdDOX-Co is the causative gene of the columnar phenotype (Okada et al., 2016; Wolters et al., 2013). The retrotransposon insertion causes the ectopic expression of MdDOX-Co in aerial organs, and MdDOX-Co degrades the precursors of bioactive gibberellins, resulting in a change in tree morphology due to a deficiency in bioactive gibberellins (Okada et al., 2020; Watanabe et al., 2021). A DNA marker developed from the inserted retrotransposon allows breeders to conduct marker-assisted selection that enables accurate selection of columnar plants for breeding (Okada et al., 2016).
The crossbreeding of columnar apples is challenging because it had to start from the original mutant ‘McIntosh Wijcik’, for which the fruits do not meet Japanese consumer preferences because of their soft flesh, low storability, low sugar content, and high acidity. To improve fruit quality attributes, NARO’s strategy was to perform pseudo backcrossing. Columnar apples and Japanese cultivars which satisfied Japanese consumer preferences were crossed to obtain seedlings. Subsequently, once promising selections were obtained, crosses were performed to obtain the next generation. As a result of more than thirty years of breeding efforts to date, a selection named Morioka 74, which is a great-great-grandchild of ‘McIntosh Wijcik’, was selected to be released as the first columnar shape cultivar from NARO. However, the disadvantage of Morioka 74 is that it has an easy-to-soften characteristic at room temperature the same as ‘McIntosh Wijcik’. Therefore, efforts should be made to improve the storability of new columnar apple cultivars.
Storability is measured as the loss of firmness and/or degree of the flesh mealiness under certain storage conditions. Quantitative trait locus (QTL) analysis, a genome-wide association study, and pedigree-based analysis (PBA) suggested that MdPG1 is the gene most contributing to the loss of firmness after two months of cold storage (Bink et al., 2014; Chagné et al., 2014; Costa et al., 2010) or mealiness after one month at 20°C storage (Moriya et al., 2017a). Gene expression analysis revealed that the expression level of MdPG1 is correlated with the loss of firmness under 24°C storage (Wakasa et al., 2006). MdPG1 down-regulated ‘Royal Gala’ confirmed the MdPG1 role in fruit softening resulted from reducing cellular adhesion (Atkinson et al., 2012). Although the responsible amino acid changes and/or promoter variations in MdPG1 remain undetermined, the Md-PG1SSR10kd simple sequence repeat (SSR) marker defines the three MdPG1 alleles (Longhi et al., 2013). Of the storability traits, the relationship between the marker genotype and mealiness was analysed in detail. Among the six genotypes defined by Md-PG1SSR10kd, MdPG1-2 homozygotes (2/2 genotype) showed the lowest mealiness, followed by 1/2, 1/1, 2/3, 1/3, and 3/3 (Moriya et al., 2017a). All apple cultivars harbouring the MdPG1-2/2 genotype, except ‘Shinano Dolce’, exhibited no mealiness (Moriya et al., 2017a). In contrast, QTL analysis and PBA have shown that MdPG1 is responsible for the crispness as measured by acoustic indices (Di Guardo et al., 2017; Longhi et al., 2012, 2013). For crispness, MdPG1-3 confers unfavourable non-crispy flesh with a dosage effect, whereas MdPG1-1 and -2 alleles confer good, crispy flesh, and the effects of MdPG1-1 and -2 alleles are not clearly different (Longhi et al., 2012, 2013). Given that MdPG1 modulates the cell wall characteristics, MdPG1, expressed in ripe apples, affects multiple flesh-related traits, including storability and texture (Atkinson et al., 2012). From a breeding perspective, the MdPG1-2 allele provides the best genetic performance for improved storability and flesh texture, whereas the MdPG1-3 allele provides the worst performance in any allele combination.
Because MdPG1 is also located on Chr 10 and ‘McIntosh’ is a homozygote of the MdPG1-3 allele (Moriya et al., 2017a), the linkage of Co and MdPG1 could induce linkage drag and hamper improvements in columnar apples, especially in terms of storability and flesh texture. In the present study, we aimed to demonstrate the existence of this linkage drag by evaluating current and historical columnar breeding materials. We then selected genetically elite plants using large-scale marker-assisted selection, which targeted individuals harbouring a favourable haplotype, leading to a columnar tree shape, high storability, and good flesh texture.
The mRNA sequence of MdPG1 (accession #L27743) and the genomic sequence of MdDOX-Co (69,147–70,777 bp; accession #LC050645) were downloaded from the National Center for Biotechnology Information (NCBI; https://blast.ncbi.nlm.nih.gov/). Nucleotide sequences were searched against published apple genomes in the Genome Database for Rosaceae (Jung et al., 2019) using the BLASTN program.
Plant materialTo evaluate linkage drag (hereafter referred to as the pilot study), current and historical breeding materials, including four cultivars, 38 selections, and 247 columnar and 46 normal individuals composed of 20 F1 populations, were used to investigate whether recombination between Co and MdPG1 occurred (Table S1; Fig. S1). The tree shape was visually assessed as columnar or normal using two-year-old or older grafted trees, or five-year-old or older self-rooted trees. Trees having shoots with thick and short internodes and less branching were scored as columnar. Genomic DNA was isolated from young leaves using an automatic DNA extraction robot, PI-50α (Kurabo, Osaka, Japan), as previously described (Moriya et al., 2009).
For marker-assisted selection, crosses were performed in each May of 2020 to 2022. The resulting seeds were sown in Jiffy-7 peat pellets (42 mm) in January and February of the following year. Plants were grown in 8 rows × 12 columns placed in plastic containers, which mimics the 96-well PCR plate design. Leaf sampling was conducted when plants reached the second true leaf stage. Approximately 2 mm2 of the tip of each young leaf was collected and placed in each well of a 96-well PCR plate, corresponding to the coordinates in the plastic container. The collected leaves were stored at −30°C or −80°C until further use. DNA was extracted using a one-step crude extraction method (Thomson and Henry, 1995) optimised by Moriya et al. (2017b). Briefly, 20 μL of template preparation solution [100 mM Tris-HCl (pH 9.5), 1 M KCl, 10 mM EDTA (pH 8.0)] was added to each well, followed by heating to 95°C for 10 min using a thermal cycler GeneAmp PCR System 9700 (Thermo Fisher Scientific, Waltham, MA, USA). Samples were diluted with 120 μL of distilled water, and 1 μL of the diluted solution was immediately used as the PCR template.
Evaluation of Co and MdPG1genotypesIn the pilot study, a Co-linked DNA marker SCAR682 (Tian et al., 2005; Table S2) located near MdPG1 on the opposite side of Co (26.84 Mb on the GDDH13 genome; Fig. S2) was first adopted based on the literature (Moriya et al., 2009) on previously tested materials or detected in newly included materials to determine whether they underwent chromosomal recombination between Co and SCAR682. PCR was performed as described by Tian et al. (2005). The Md-PG1SSR10kd SSR marker (27.28 Mb on GDDH13 genome; Longhi et al., 2013; Table S2; Fig. S2) was used to identify the genotypes of MdPG1. PCR and detection were performed as described by Longhi et al. (2012), except that a CEQ8000 capillary sequencer (SCIEX, Framingham, MA, USA) and corresponding fluorescent dyes (D2, D3, and D4) were used. Moreover, the SSR marker CH03d11 (27.37 Mb on the GDDH13 genome; Liebhard et al., 2002; Table S2; Fig. S2) was also tested or adopted from the literature (Moriya et al., 2009) to confirm recombination if required. The genotypes of Co were determined based on tree shape. Columnar and normal plants were considered as Co/co and co/co, respectively.
In the marker-assisted selection experiment, MdPG1 and Co genotypes were evaluated using a multiplex PCR strategy. The MdPG1 genotype was evaluated using a primer set designed from the promoter sequence of MdPG1, which specifically amplified the MdPG1-3 allele (Table 1; Costa et al., 2010). The forward primer sequence was modified from the experiment in 2023 to improve specificity. The MdPG1-1 and -2 alleles were not tested. The genotypes of the Co locus were evaluated using the aforementioned primer set (Okada et al., 2016; Table 1). Multiplex PCR with these two primer sets was performed using Go Taq G2 Hot Start Master Mix (Promega, Madison, MI, USA) and the final primer concentration, as listed in Table 1, in a 10 μL reaction mixture. The PCR conditions included an initial denaturation at 94°C for 2 min, followed by 5 cycles of touch-down PCR: denaturation at 94°C for 30 s, annealing at 60°C for 90 s (−0.5°C/cycle), and extension at 72°C for 50 s, followed by 32 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 45 s, and extension at 72°C for 40 s, and a final extension at 72°C for 4 min. The amplified PCR products were separated by electrophoresis on a 2% agarose gel, stained with ethidium bromide, and visualised under ultraviolet light. Individuals with MdPG1-3-negative and Co-positive results were selected.
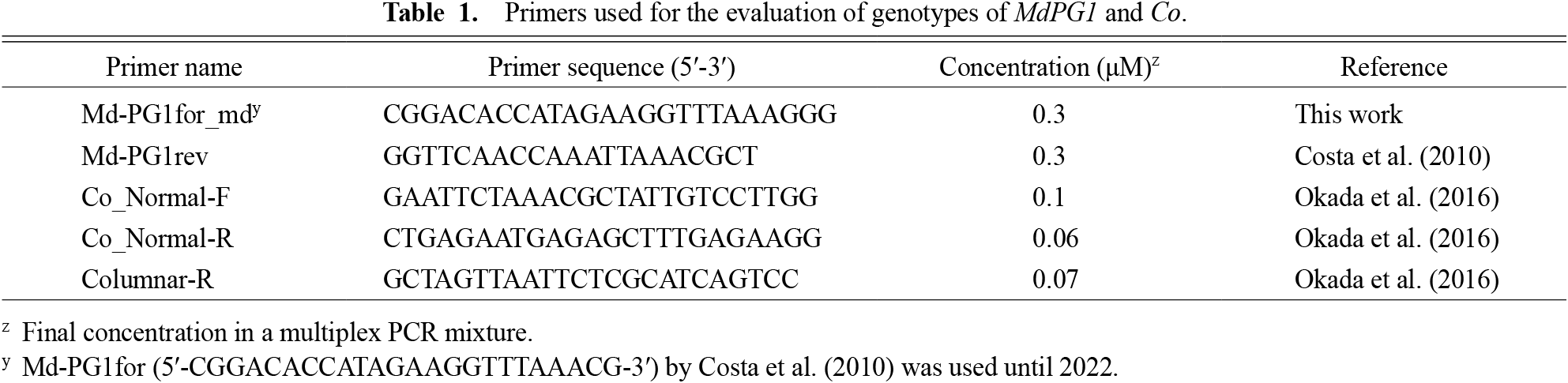
Primers used for the evaluation of genotypes of MdPG1 and Co.
The BLASTN search identified MdDOX-Co and MdPG1 sequences on Chr 10 in several published apple genome sequences (Daccord et al., 2017; Khan et al., 2022; Sun et al., 2020; Zhang et al., 2019; Table 2). Because Chr 10A of the ‘Gala’ diploid genome contained two MdDOX-Co and two MdPG1 sequences, the closest pair was used to calculate physical distance. The distance ranged from 456 kb (Chr 10B of ‘Gala’) to 750 kb (Chr 10A of M. sylvestris) and averaged 633 kb. Because previous studies determined the physical length and genetic distance of the entire Chr 10 to be 41,762 kb (Daccord et al., 2017) and 81.3 cM (Di Pierro et al., 2016), respectively, 633 kb was estimated to be a 1.23 centimorgan (cM).

Physical distances between MdPG1 and MdDOX-Co in apple genomes.
‘McIntosh Wijcik’ had the MdPG1-3/3 genotype, indicating that the columnar tree shape-inducing (Co) and low storability-inducing (MdPG1-3) alleles were linked in the coupling phase (the Co–MdPG1-3 haplotype) (Table 3; Fig. 1). SCAR682 was detected in all columnar cultivars/selections presented in Table 3 and Figure S1, except 5-263 and 8-937, indicating that all materials except two retained the original Co–MdPG1-3 haplotype (Table 3). 8-937 had the MdPG1-2/3 genotype; however, the MdPG1-2 allele was transmitted from the normal tree-shaped cultivar ‘Shinano Gold’ (MdPG1-2/2), indicating that 8-937 retained the Co–MdPG1-3 haplotype (Fig. 1). Regarding 5-263, the MdPG1 genotype could not confirm any recombination event due to its homozygous genotype (MdPG1-3/3); however, the genotype of CH03d11 (118 bp/118 bp) adopted from Moriya et al. (2009) suggested recombination between Co and CH03d11 because the Co-coupling allele of CH03d11 was 177 bp (Moriya et al., 2009; Fig. 1). Therefore, the Co-located Chr 10 of 5-263 underwent recombination between Co and MdPG1, but it was a newly emerged Co–MdPG1-3 haplotype.

Haplotype estimation and DNA marker genotypes of columnar/normal apples.

Graphical genotypes of chromosome 10 in key plant materials in the pilot study. Co-associated alleles are shown in dark grey, other alleles are shown in light grey, and undetermined alleles are shown in medium grey.
Of the 247 individuals from 20 historical breeding populations (Table S1), three individuals (6-5579, 6-5657 and #858 from the PS05, PS11, and PS20 populations, respectively) showed recombination between Co and SCAR682 (Fig. 1). Of the three individuals, 6-5579 and #858 confirmed recombination between Co and MdPG1, resulting in the formation of a novel and beneficial haplotype carrying both Co and MdPG1-2 alleles (Co–MdPG1-2 haplotype; Fig. 1). Nevertheless, for the remaining 6-5657 detected recombination events between CH03d11 and SCAR682, the exact region of recombination could not be confirmed because of the homozygosity of the MdPG1 (MdPG1-3/3) locus in its columnar parent 8S-60-74. Consequently, three (5-263, 6-5579, and #858) of the 335 materials, including columnar cultivars, selections, and segregating populations used in the pilot study (approximately 0.9%) showed recombination between Co and MdPG1; one (6-5657) could not be determined, and the remaining 331 retained the original haplotype. Of the three recombinants, #858 and 6-5579 carried the Co–MdPG1-2 haplotype.
Large-scale selection of a rare individual carrying the Co–MdPG1-2 haplotypeFive crosses were made between normal tree-shaped cultivars with the MdPG1-2/2 or 1/2 genotypes and columnar apples with the Co–MdPG1-3/co–MdPG1-2 diplotype (Table 4). The number of seeds sown ranged from 809 to 12,290 and the number of individuals evaluated using DNA markers ranged from 585 to 9,680. The reason for the reduction in the number of individuals evaluated from the sown seeds was the occurrence of non-germinating seeds and a pale green lethal disorder. Marker detection resulted in the identification of 2–57 individuals showing MdPG1-3-negative and Co-positive individuals (Table 4; Fig. 2). Because the normal tree-shaped cultivars used as parents did not harbour the MdPG1-3 allele (Table 3), individuals showing MdPG1-3-negative and Co-positive results indicated that they carried the Co–MdPG1-2 haplotype. Subsequently, their MdPG1 genotypes were confirmed by the Md-PG1SSR10kd marker and selected. The proportion of selected individuals varied between 0.10%–1.54% for the respective crosses, and 0.53% for the total.

Large-scale selection of individuals carrying the chromosome with Co and MdPG1-2 in the coupling phase (the Co–MdPG1-2 haplotype).

Genotyping of Co and MdPG1 by multiplex PCR. F1 individuals from the ‘Kitaro’ × 8-7573 cross (LS05 population) are shown. Genomic DNA was extracted according to Thomson and Henry (1995). The sample marked S in the decision row is an individual carrying a chromosome with both Co and MdPG1-2 in the coupling phase (Co–MdPG1-2 haplotype). 8-818 is a normal tree-shaped breeding selection harbouring the MdPG1-1/2 genotype. S: selected.
In the present study, we demonstrated a strong linkage between Co and MdPG1, which could hamper storability and flesh texture improvement of columnar apples. A large-scale marker-assisted selection experiment succeeded in selecting 80 individuals that carried a favourable haplotype (Co–MdPG1-2), which is likely to have the potential to dramatically improve the storability of columnar apples.
To develop columnar cultivars with long-term storability and favourable flesh characteristics, two bottlenecks were identified in the present study. The first is the strong linkage between Co and MdPG1, which hinders the recombination between Co and MdPG1-3. Apples with the MdPG1-3 allele have mealy flesh when stored at room temperature (Moriya et al., 2017a). A high frequency (98.8%) of plant materials retained the original haplotype (the Co–MdPG1-3 haplotype in columnar materials) in the pilot study, suggesting that the improvement in storability was disturbed by this linkage drag in columnar apple breeding. Therefore, the Co–MdPG1-3 haplotype should be removed from breeding programs and a Co–MdPG1-2 haplotype that potentially can achieve good storability should be constructed and introduced as a promising breeding material. Because the computationally estimated genetic distance between Co and MdPG1 is 1.23 cM, recombination in this region can occur in 1.23% of the gametes. Using columnar parents that harbour the Co–MdPG1-3/co–MdPG1-2 diplotype, such as the 7-4151 selection (Table 3), four types of gametes are expected: 49.4% each of the Co–MdPG1-3 and co–MdPG1-2 haplotypes (original haplotypes) and 0.6% each of the Co–MdPG1-2 and co–MdPG1-3 haplotypes (recombinant haplotypes). Only Co–MdPG1-2 was valuable for columnar apple breeding.
The result of the pilot study, which identified 0.9% of the individuals carrying the recombinant haplotype on the chromosome from the columnar parent, was close to the value estimated in our calculation (1.23%) using in silico analysis. In terms of large-scale selection, only the number of the target haplotype (Co–MdPG1-2) was counted, excluding other haplotypes (Co–MdPG1-3, co–MdPG1-2, and co–MdPG1-2) due to the laborious nature of the task, meaning that the selection rate (0.53%) was lower than the actual recombination value. To estimate the true recombination value between Co and MdPG1, it is likely appropriate to assume that an additional 0.53% of the recombinant haplotype (co–MdPG1-2) was included in the remaining individuals. As a result, the estimated true recombinant frequency for our large-scale selection was 1.06%, which is almost consistent with the genetic distance (1.23%) estimated using in silico analysis.
The NARO apple breeding program normally plants 1,200–1,500 new apple seedlings in an experimental orchard every year. If we plan to obtain 10 columnar apple seedlings that carry the Co–MdPG1-2 haplotype without any selection before planting, the required population size will be approximately 1,700 individuals, meaning that all resources, such as an orchard and labour for one year, must be used only for this objective, which is unrealistic in practical breeding. Therefore, the emergence of recombination and selection of recombinant gametes are important. Large-scale marker-assisted selection provides a solution for selecting seedlings carrying the Co–MdPG1-2 haplotype as early as possible. In this study, 18,000 individuals were developed, and 80 individuals were selected. This means that marker-assisted selection saved land, labour, and related costs equal to more than 10 years of breeding efforts. In addition, the crude extraction protocol used in this study is cost-effective and easy to perform. The components of the extraction buffer included only generally available and inexpensive chemicals. The plant growing system had a consistent placement in the 96-well PCR plate format, making it possible to skip the plant marking or numbering steps and enable leaf sampling from 96 plants in 15 min. Although agarose gel electrophoresis was used to detect PCR products in this study, more high-throughput systems, such as the KASPTM genotyping assay (Baumgartner et al., 2016) may be considered.
The second bottleneck is the MdPG1 genotype of the columnar shaped breeding material. Designing the Co–MdPG1-2 haplotype, using columnar apples with the MdPG1-2/3 genotype is a prerequisite. The 5-263 selection resulted in the formation of a novel Co–MdPG1-3 haplotype because the MdPG1 genotype of its columnar parent 8H-6-4 was MdPG1-3/3 (Table 3). Fifteen columnar cultivar/selections were identified as MdPG1-2/3 (Table 3) and were used as crossing parents to obtain the Co–MdPG1-2 haplotype in a large-scale marker-assisted selection in the present study. Because ‘McIntosh’ shows many unfavourable fruit characteristics unacceptable in the Japanese market, fruit quality improvement is a primary objective in the columnar apple breeding. Because the genetic potential for improvement of fruit quality is elevated in each successive generation from ‘McIntosh Wijcik’, the latest selections such as 7-4151 and 8-7573 (great-great-grandchildren of ‘McIntosh’; Table 3; Fig. S1) are promising parents for obtaining high-fruit quality columnar apples in the subsequent generation. These selections show higher sugar content and lower acid compared to those from previous generations.
In a large-scale marker-assisted selection experiment, PCR results were stable in experiments conducted in 2021 (experiments for crossing in 2020), but sometimes became unstable in 2022 (experiments for crossings in 2021), probably because of the different primer suppliers from 2021: the MdPG1-3 allele was amplified from all samples tested in the same batch (data not shown). Repeat PCR using the same DNA template yielded appropriate results in almost all cases. Modification of the forward primer sequence in 2023 (crossing experiments in 2022) improved the PCR results, likely because of the increased specificity of the primer.
Although 80 individuals carrying the Co–MdPG1-2 haplotype were selected, we could not determine the number of plants required to obtain promising seedlings that fulfilled our selection criteria for fruit quality. Although Co–MdPG1-2 haplotype carriers (6-5579 and #858 found in historical breeding populations) were identified, according to our breeding records, they were culled more than 10 years ago; 6-5579 were culled because of poor colouration of the fruit skin. We could not track the sensory evaluation results of #858 because it was the only plant ID used for genetic mapping, and we could not match the corresponding plant ID in our breeding orchard. This result clearly suggests that even individuals with the ideal haplotype cannot always meet the standards for other fruit qualities, such as fruit colour, acidity, and sweetness, meaning that the greater the number of individuals developed, the greater the chance for the selection of promising seedlings. Therefore, a large number of individuals should be developed and selected initially.
In conclusion, 80 individuals selected by large-scale marker-assisted selection experiments, which were expected to show a columnar shape with improved storability and flesh texture, were grown in our experimental orchard. Although sensory evaluation of the fruit is necessary for the selection procedure, advanced columnar materials with superior storability and flesh texture will be obtained in the next few years. Once such material is established, large-scale experiments to select recombinant haplotypes will no longer be necessary. The strong linkage between the two genes guarantees the transmission of the Co–MdPG1-2 haplotype to the offspring. Superior columnar cultivars can be game changers in columnar apple breeding and provide valuable material for the development of highly profitable and labour-saving apple cultivation systems using columnar apple cultivars.
We appreciate the technical assistance provided by the Genome Breeding Support Office of the Institute of Crop Science at NARO (NICS). The authors declare no conflicts of interest.