2017 Volume 42 Issue 3 Pages 105-111
 J-STAGE : jpestics 43 1 pp.55-55
J-STAGE : jpestics 43 1 pp.55-55 J-STAGE : jpestics 43 3 pp.224-224
J-STAGE : jpestics 43 3 pp.224-224 Browse “Advance online publication” version
Browse “Advance online publication” version2017 Volume 42 Issue 3 Pages 105-111
 J-STAGE : jpestics 43 1 pp.55-55
J-STAGE : jpestics 43 1 pp.55-55 J-STAGE : jpestics 43 3 pp.224-224
J-STAGE : jpestics 43 3 pp.224-224 Browse “Advance online publication” version
Browse “Advance online publication” versionFourteen compounds screened from 5 million compounds in silico were submitted to bioassay to find brassinolide (BL) agonists/antagonists against Arabidopsis thaliana. Of these, two N-benzoyl-N′-phenylpiperazine (NBNPP)-type compounds showed antagonistic activity; however, none showed agonistic activity against A. thaliana. The substituents at the benzoyl moiety of NBNPP were changed to OH groups to derive N-(3,4-dihydroxybenzoyl)-N′-(4-butanoyl-2-fluorophenyl)pyrazine, which was named NSBR1. NSBR1 was rationally designed based on docking simulations and molecular dynamics. NSBR1 significantly suppressed the gene expression of CPD and BR6-ox2, which are known as marker genes for the action of BL. This novel NSBR1 was also effective in the rice lamina inclination assay (RLIA), and the activity in terms of the 50% effective dose (ED50) was determined as 0.79 nmol/plant from the dose–response curve for RLIA.
Brassinolide (BL; 1 in Fig. 1) is a plant steroid hormone; its chemical structure was identified in 1979.1) BL and its related compounds are classified as brassinosteroids (BRs). BRs are present throughout the plant kingdom at low concentrations and play an important role in plant growth and development.2) To date, more than 70 BRs have been identified in nature, and some BRs have been chemically synthesized.3–6) BRs elicit a broad spectrum of physiological and morphological responses in plants: stem elongation, leaf bending and epinasty, induction of ethylene biosynthesis, activation of proton pumps, biosynthesis of nucleic acid and proteins, regulation of carbohydrate assimilation and allocation, and activation of photosynthesis.7) The BR receptor first identified by Wang et al. was named BR-insensitive receptor kinase 1 (BRI1),8) and the three-dimensional structure of BRI1 was solved by using X-ray crystal structure analysis.9,10) In the meantime, it was reported that somatic embryogenesis receptor kinase 3 (SERK3), which is known as BRI1-associated receptor kinase 1 (BAK1), is required for the signal transduction of BRs.11) A few years ago, the crystal structure of the BRI1-BL-SERK1 complex of Arabidopsis thaliana was first published.12)
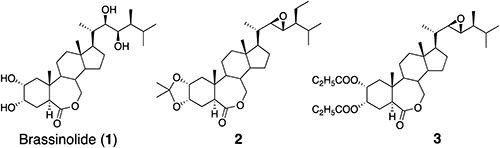
Since BRs can protect plants from various abiotic stresses, induced by high salt, high temperatures, heavy metals, and drought7); and biotic stress elicited by bacteria, viruses, fungi, parasites, and insects13); compounds that mimic the action of BRs can be useful agrochemicals. The defensive effects of BRs against herbicides14) and prospective medical applications are also attractive.15) In the late 20th century, there were several reports suggesting that BRs would be put into practical use in the near future.16) Some pesticide companies have worked toward the practical use of BRs such as BL (1), 24-epiBL, and (22S,23S)-28-homoBL; however, the extent of the effectiveness and the stability of results decreased.7) Then, modified BL analogs, such as 2 and 3 (TS303), were selected as other candidates for practical use.16) TS303 and TNZ303 liquid containing TS303 and n-propyl dihydrojasmonate significantly increased crop yields. The spray treatment of rice seeds with TNZ303 increased the yield. In further studies, this spray treatment was applied to wheat, potatoes, and sugar beets in official tests in Russia and Poland.16) However, only a few scientific papers regarding the application of BRs have been published during the last two decades. BRs can be used for plant protection and yield promotion17,18); however, the basic structure (steroid skeleton) of BRs may interfere with the practical use of BRs in agriculture.
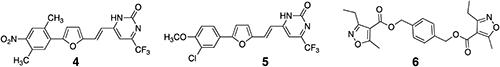
The conversion of steroids to non-steroids is very attractive because the discovery of nonsteroidal insect molting hormone agonists opened the way for the development of a class of novel insecticides.19–22) Approximately two decades ago, nonsteroidal BL-like compounds were synthesized, and their activity was measured by rice lamina inclination assay (RLIA).23) However, since the activity was implausible, Takimoto synthesized this reported compound24) and found that it was inactive against cress treated with a BL biosynthesis inhibitor, brassinazole (Brz).25) We previously searched for nonsteroidal BL-like compounds using in silico screening and subjected the screened compounds to RLIA. Unfortunately, no compound was found to be an agonist in RLIA26); however, three compounds (4–6, Fig. 2) were found to be antagonists. The final goal of our study is to find a novel nonsteroidal BL agonist. In this study, the compounds that were found to be BL antagonists were rationally converted to agonists using in silico techniques, such as docking simulation and molecular dynamics (MD). The designed compounds were synthesized and submitted to bioassay to detect BL activity. The expression of marker genes that respond to BL treatment was examined in Arabidopsis.
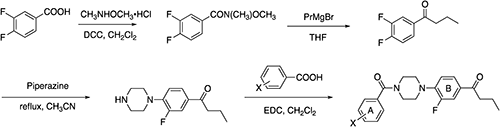
Compounds 4 and 6–8 (Fig. 2 and Fig. 3) are from our stock sample. Novel N-benzoyl-N′-phenylpiperazine (NBNPP) analogs 9–1226) were chemically synthesized from piperazine and substituted benzoic acids in accordance with synthetic Scheme 1. Structures of newly synthesized compounds were identified by 1H and 13C NMR (BRUKER AVANCE 400) and high-resolution mass spectra (HRMS). HRMS was recorded on a Thermo Fisher Scientific EXACTIVE spectrometer at the Department of Synthetic Chemistry and Biological Chemistry of Kyoto University. Purification by chromatography was performed using Biotage SNAP Ultra (Biotage Japan, Tokyo). The detail of the chemical synthesis is shown in the supplemental file (Supplementary Material).
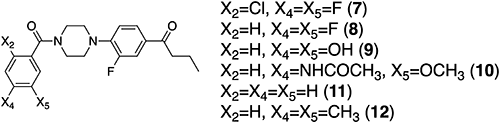
Seeds of A. thaliana Col1-0 were immersed in 70% EtOH containing 1% Triton X-100 for 30 min and then transferred to 99% EtOH and stirred for 1 min. Seeds were put on sterilized filter paper to remove the EtOH. Sterilized seeds were planted in a 1/2 MS medium containing 0.9% agar, 1.5% sucrose, and test compounds and grown at 4°C under dark conditions. After standing for 2–3 days, the seeds were held for 4 hr under light at 22°C (vernalization). After growing for 7 days at 22°C under dark conditions, the length of the hypocotyls was measured. The hypocotyl length was measured using ImageJ pictures of the hypocotyls. Test compounds were added in the medium as a DMSO solution (less than 1% DMSO). Parts (50 mg) of the plants were stored in 1 mL RLT buffer (RNeasy Plant Mini Kit; Qiagen, Heiden, Germany) containing 10 µL of mercaptoethanol at −80°C. The frozen plant pieces were ground by mortar and pestle and subjected to RT-PCR using a TaKaRa PrimeScript RT Reagent Kit. The solution of the RT-PCR reaction (25 µL) consisted of SYBR Premix Ex Taq II (12.5 µL), sterilized water (7.3 µL), forward and reverse primers (0.1 µL), and cDNA (2.5 µL).
2.2. Rice lamina inclination assayA rice lamina inclination assay using seedlings of dwarf rice Oryza sativa cv. Tan-ginbozu was used to measure the BL-like activity.3) Tan-ginbozu seeds were sterilized with a 2% aqueous NaClO solution and germinated by soaking in the sterilized water for three days under light at 30°C. The sprouted rice seeds were transplanted into the 1% agar medium and cultured for three days under light at 30°C. The EtOH solution of the test compound was applied to the second lamina of the rice shoot after the application of 0.5 µL of indole-3-acetic acid (50 mM in EtOH). Treatments with 1 µL of EtOH and BL solution (1 µM) were used as negative and positive controls, respectively.
3. MD simulationThe method for MD and the binding free energy calculation of the ligand–receptor complex have been previously reported.27) MD simulation was carried out using an AMBER 14 MD package28) and AMBER Tools 15. An AMBER force field 14SB29,30) was applied for the protein and gaff31) of the ligand molecules. Free energy was calculated by the MM-PBSA (Molecular Mechanics Poisson-Boltzmann Surface Area)32) as ΔGmmpbsa, and the ligand conformer entropy was calculated using the Freeform module of SZYBKI (Ver. 1.8.0.1; OpenEye: Santa Fe, USA). The binding energy in terms of ΔGbind is calibrated as ΔGmmpbsa−TΔSligand. Conformation ensembles for each ligand molecule were generated by OMEGA2 (Ver. 2.5.1.4). The receptor protein complex model was constructed from two crystal structures (PDB: 4LSX and PDB: 4LSA) by removing the ligand molecule BL. To construct the BRI1/SERK1 complex model, chain A (BRI1) of 4LSX was replaced with 4LSA, and chain C of 4LSX was used as the SERK1 model. Docking simulation was performed using MAKE_RECEPTOR and FRED modules of OEDocking (Ver. 3.2.0.2; OpenEye).33)
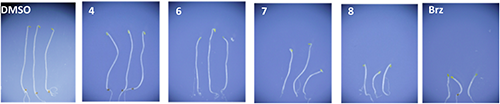
Fourteen compounds, including 7 and 8, which were screened in our previous study,26) were subjected to bioassay using A. thaliana. We were unable to assay compound 5 due to insufficient quantities. Compounds 4 and 6 had no effect in the Arabidopsis hypocotyl elongation assay; however, compounds 7 and 8 with an NBNPP basic structure significantly suppressed hypocotyl elongation at 100 µM, as shown in Fig. 4. This effect was similar to that shown with Brz treatment at 1 µM. Other screened compounds26) were inactive in this Arabidopsis hypocotyl elongation assay.
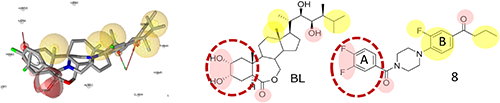
The superposition of structures of BL and 8 on the pharmacophore model used in our previous in silico screening is shown in Fig. 5.26) The pink spheres surrounding F atoms of the A ring of 8 correspond to the hydrogen bonds formed between hydroxyl groups of the steroid A rings of BL and SERK1. The carbonyl group between the benzene ring and the piperazine ring of compound 8 corresponds to the carbonyl group on the steroid B ring (ε lactone ring) of BL. Another carbonyl oxygen close to the B ring of compound 8 corresponds to the oxygen of the OH group at C-22 of the side chain of BL. Yellow spheres surrounding C-18 and C-21 are hydrophobic features that correspond to the fluorophenyl moiety of 8. The other sphere surrounding the terminal n-propyl moiety of 8 matches that of the terminal isopropyl group of the side chain of BL.
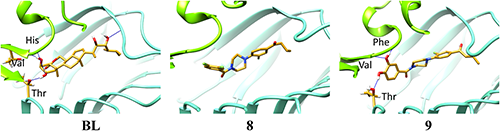
In order to compare the interactions between receptor proteins and ligand molecules, BL and 8, respectively, we performed a docking simulation using FRED (OpenEye) and calculated the binding free energy using MD/MM-PBSA.32) As shown in Fig. 6, hydrogen bonds (HBs) between OHs of the BL A-ring and SERK1 were detected (left figure of Fig. 6), while no hydrogen bond (HB) was formulated for the antagonistic compound 8 (middle figure of Fig. 6). In the original crystal structure of the BRI-BL-SERK1 complex (PDB: 4LSX), these two OH groups are important for the interaction between BL and SERK1.12) Thus, we replaced F atoms of 8 with OH to give 9 so as to mimic the A-ring moiety of BL and docked 9 to the receptor. As predicted, HBs were formulated between OH and SERK1 for 9 after MD simulation. Phe and His residues of SERK1 participate in HB formation with 2,3-(OH)2 of the A ring of BL in the original crystal structure (4LSX); however, different amino acid residues (His, Val, Thr) participate in HB formation in the newly constructed BRI1/SERK1 model after MD. In the docking model for 9, three amino acids (Phe, Val, Thr) participate in HB formation, which was slightly different from that for BL; however, no HBs were observed for 8. Interestingly, Thr residue of BRI1 formulated HB in both BL and 9 complex. Among these amino acid residues formulating HBs, Phe and His are located vicinally. ΔG values of ligand–receptor binding for candidate compounds 7–12 using MM-PBSA and Freeform (OpenEye) are listed in Table 1. Even though the ΔG values of all compounds are not negative, compound 9 has the lowest ΔG value of the six NBNPPs (Table 1).
| Compound | ΔG (kcal/mol) | Compound | ΔG (kcal/mol) |
|---|---|---|---|
| 7 | 8.33 | 10 | 17.14 |
| 8 | 9.85 | 11 | 11.13 |
| 9 | 2.18 | 12 | 7.30 |
a) ΔG: BL −5.81 kcal/mol; CS −5.30 kcal/mol
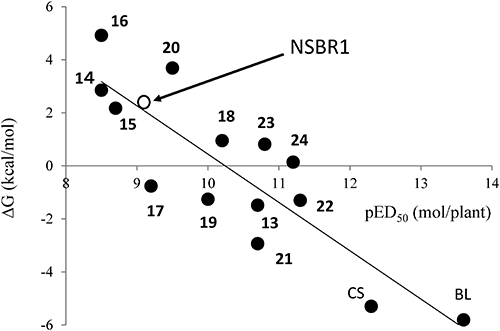
Previously, we had synthesized CS analogs (13–16; Fig. 7) with various hydroxyalkyl chains and measured their activity in order to discuss the effect of conformation on the activity.5) We recently synthesized CS analogs (17–24; Fig. 7) with various alkyl chains and measured their activity in order to discuss the structure–activity relationship.34) In both studies, the activity was quantitatively evaluated as pED50 (ED50=50% effective dose) by RLIA. We also calculated the ΔG values of CS agonists (Fig. 7) as well as those of BL and CS, and we found the linear correlation between ΔG and pED50.34) In this calculation condition, the ΔG values of some compounds were positive, which seems to be theoretically unacceptable; however, these calculated values correlated nicely with the in vivo activity, as shown in Fig. 8. As shown in Fig. 8, compound 9 (ΔG=2.18 kcal/mol) is predicted to be active from the linear correlation between ΔG and pED50 measured for steroidal compounds,34) although 4 other compounds (7, 8, 10–12) were thought to be inactive due to the high ΔG (7.30–17.14 kcal/mol).
Since compound 9 is proposed to be a BL agonist, its biological activity was examined using Arabidopsis hypocotyl elongation assay. As shown in Fig. 9, the Arabidopsis hypocotyl elongation suppressed by Brz (1 µM) treatment was significantly recovered by treatment with 9 as well as BL.

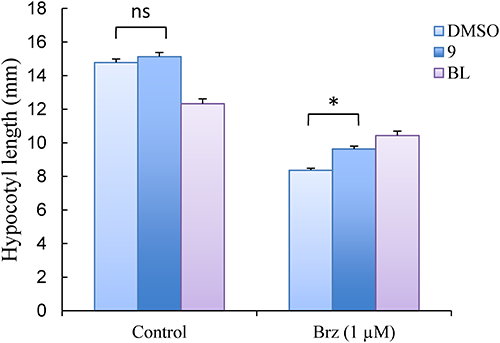
Among the newly synthesized NBNPPs (9–12), 9 significantly elongated the Arabidopsis hypocotyl treated with Brz. Therefore, we assessed the effects of 9 on BR-responsive gene expression. Seven-day-old wild-type Arabidopsis plants grown in half MS agar-solidified media treated with or without 9 were used for quantitative real-time PCR (qPCR) analysis. The concentrations of 9 used in this assay were 10 and 30 µM, at which, importantly, it induces BL-like phenotypes in Arabidopsis. We used 100 nM of BL and 3 µM of Brz25) as a control to determine the effect of 9 on BR-responsive marker genes. In this study, we chose BL biosynthesis genes (CPD and BR6ox2) that are down-regulated by BL.35–37) Consequently, we found that 9 down-regulated the expression levels of these genes in a dose-dependent manner, mimicking the action of BL (Fig. 10). These results indicate that 9 is a BL agonist, activating hormonal responses through a common transcriptional module. Other compounds (10–12) had no effect on marker gene expressions in the Arabidopsis assay (data not shown).

Since 9 showed an agonist effect on Arabidopsis, these newly synthesized NBNPPs (9–12) were also evaluated against rice using RLIA (Fig. 11). Only 9 showed significant agonist activity in RLIA at 10 nmol. In further studies, we examined the dose–response relationship for 9 (Fig. 12). The ED50 of 9 was calculated as 0.79 nmol (pED50=9.1) from this dose–response curve by probit analysis.38) This activity is reasonably predicted from the relationship between ΔG and pEC50 expressed in Fig. 8.
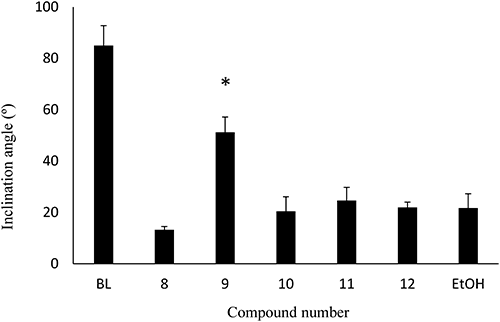
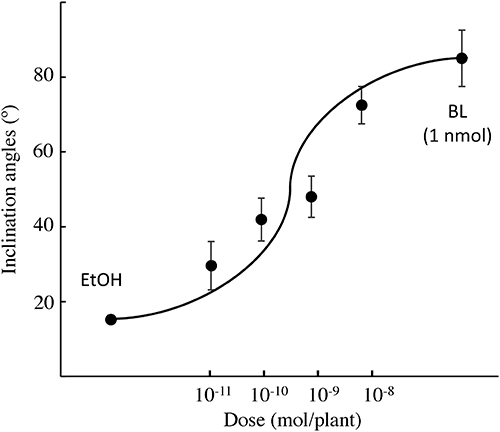
Although the potency of 9 (pED50=9.1) was weak compared to BL (pED50=13.6) and CS (pED50=12.3), it was 2–3 times more potent than other CS analogs, 21-epi-(pED50=8.5) and 21-nor-CS (pED50=8.7)5) and equipotent to 17 (pED50=9.2).34) Compound 9 was also equipotent to BL analogs with acyl side chains at the C-17 position (pED50=8.6–10.5).6) Since this new nonsteroidal compound 9 showed BL-like activity under both physiological and genomic conditions, it was named NSBR1. The binding energy in terms of the ΔG (2.13 kcal/mol) calculated by MM-PBSA was reasonable, as shown in Fig. 8. Further modification of substituents at the A and B rings of NBNPP, as well as the replacement of its piperazine and benzene rings by more appropriate moieties, is expected to enhance activity, which is now in progress. This result indicates that the MM-PBSA condition used to calculate the binding free energy (ΔG) of ecdysone agonists27) is also applicable to other ligand–receptor binding energy calculations.
Fourteen compounds in silico screened from 5 million compounds were subjected to Arabidopsis assay to find agonists/antagonists of BL. Two NBNPPs showed antagonistic activity, and one was successfully converted to an agonist, NSBR1. The structure conversion from antagonist to agonist was done rationally by considering the ligand–receptor binding energy based on MD simulation. NSBR1 also showed agonistic activity in the gene expression of Arabidopsis and RLIA.