2014 Volume 64 Issue 4 Pages 371-377
2014 Volume 64 Issue 4 Pages 371-377
Soybean (Glycine max (L.) Merr.) oil typically contains 8% α-linolenic acid that is highly unstable and easily oxidized. This property is undesirable in many food and industrial applications. Genetic strategies for reducing α-linolenic acid content would enhance the commercial value. However, genetic resources for low α-linolenic acid content are limited among natural soybean variations. Microsomal omega-3-fatty acid desaturase (FAD3) is responsible for the synthesis of α-linolenic acid in the polyunsaturated fatty acid pathway. There are four FAD3 homologs (Glyma02g39230, Glyma11g27190, Glyma14g37350 and Glyma18g06950) in the soybean genome. While non-functional alleles have been reported for Glyma02g39230 (GmFAD3-1a) and Glyma14g37350 (GmFAD3-1b), little variation is seen in Glyma18g06950 (GmFAD3-2a). We isolated seven mutant GmFAD3-2a alleles, each containing a single-nucleotide substitution, from 39,100 independent mutant lines by using targeting induced local lesions in genomes (TILLING). Analysis of GmFAD3-2a transcripts and enzyme activities revealed that one missense mutant, ‘P1-A9’, contains a non-functional allele of GmFAD3-2a. By combining three non-functional alleles (GmFAD3-1a, GmFAD3-1b, and GmFAD3-2a), we generated soybean lines containing <2% α-linolenic acid in their seeds. The reverse-genetics-based development of novel mutant alleles in the fatty acid metabolic pathway will allow the improvement of soybean with better oil quality through conventional breeding.
Fatty acid composition is an important factor in determining the quality of soybean (Glycine max (L.) Merr.) oil. Five fatty acids are predominant in soybean: palmitic, stearic, oleic, linoleic, and α-linolenic acids. Of these, α-linolenic acid is an unstable component responsible for unpleasant odor and reduced shelf life, characteristics that are associated with poor oil quality (Liu and White 1992, Shen et al. 1997). To overcome these disadvantages, soybean oil is usually hydrogenated to reduce the α-linolenic acid content (Kinney 1996, Yusem and Pintauro 1992), but hydrogenation generates trans-fatty acids that increase the risk of obesity and coronary heart disease (Mozaffarian et al. 2006, Zaloga et al. 2006). Therefore, a considerable reduction in α-linolenic acid content is a desired breeding objective, to produce oxidatively stable soybean oil without the generation of trans-fatty acids.
In developing seeds, α-linolenic acid is synthesized by omega-3-fatty acid desaturase (FAD3) in the polyunsaturated fatty acid biosynthesis pathway (Yadav et al. 1993). The soybean genome contains four FAD3 genes, of which at least three (GmFAD3-1a, GmFAD3-1b, and GmFAD3-2a) appear to be functional because their transcripts accumulate in developing seeds (Anai et al. 2005, Bilyeu et al. 2003). Thus, GmFAD3-1a, GmFAD3-1b, and GmFAD3-2a are considered to play important roles in controlling α-linolenic acid levels in developing soybean seeds and have been targeted in the development of low-α-linolenic acid soybeans. Low-α-linolenic acid soybean lines that have been reported have one or more mutations in these genes (Bilyeu et al. 2003, 2005, Chappell and Bilyeu 2006, 2007, Reinprecht et al. 2009). Some studies have associated combinations of mutations in the three genes with a 1% α-linolenic acid content (Bilyeu et al. 2006, 2011). However, soybean genetic resources for improving this trait remain limited.
We previously isolated two low-α-linolenic acid soybean mutants, ‘M24’ and ‘M5’, by phenotype screening (Rahman et al. 1996, 1998), and showed that they have non-functional alleles of GmFAD3-1a and GmFAD3-1b, which contain 1- and 19-bp deletions, respectively, in their open reading frames (ORFs) (Anai et al. 2005). The double-recessive mutant line ‘LOLL’ was developed from an ‘M24’ × ‘M5’ cross, and its α-linolenic acid content was reduced to 2.3% (Rahman et al. 1998). To achieve novel soybean germplasms with a maximum of 1% α-linolenic acid content, a mutation in GmFAD3-2a is necessary. However, no mutants of GmFAD3-2a were obtained in our previous study using phenotype-based forward genetics screening, possibly because GmFAD3-2a contributes less to α-linolenic acid production than the other two isozymes (Anai et al. 2005). Furthermore, environmental conditions might mask the phenotype of a GmFAD3-2a mutant, because temperatures during maturation strongly affect polyunsaturated fatty acid levels in soybean seeds (Dornbos and Mullen 1992, Rennie and Tanner 1989). Therefore, alternative approaches are needed to obtain novel mutant alleles of GmFAD3-2a.
TILLING (targeting induced local lesions in genomes) is a high-throughput reverse genetics technique used for identifying novel mutant alleles from mutagenized populations (McCallum et al. 2000, Olekowski et al. 1998). It can easily generate more alleles than naturally occurring variations at a target locus, and elite mutant alleles are readily available in conventional breeding programs (Anai 2012, Henikoff et al. 2004). We recently constructed a high-throughput soybean TILLING system (Anai 2012) and isolated novel alleles of genes that control maturity (Watanabe et al. 2009, 2011, Xia et al. 2012) and seed oil quality (Hoshino et al. 2010). Here, we selected GmFAD3-2a as a primary target for developing novel low α-linolenic acid alleles by TILLING. We screened 35 800 independent mutant lines and identified a null mutant allele of GmFAD3-2a. In addition, we generated soybean lines containing <2% α-linolenic acid by combining mutant alleles of GmFAD3-1a, GmFAD3-1b, and GmFAD3-2a. We show the development of this novel genetic resource for improving the α-linolenic acid content of soybean oil.
Two low-α-linolenic acid soybean mutants, ‘M24’ and ‘M5’, were previously obtained from an X-ray irradiated population of ‘Bay’ at Saga University, Saga, Japan. The double-mutant ‘LOLL’ was developed from a cross between ‘M24’ and ‘M5’ (Rahman et al. 2001). The low-α-linolenic acid mutant ‘P1-A9’ was obtained from an X-ray irradiated population of ‘Bay’ by TILLING (in this study), and two independent triple mutants, ‘LOLL-A9-1’ and ‘LOLL-A9-2’ were developed from 109 F2 progeny of a cross between ‘P1-A9’ and ‘LOLL’. These mutant lines and ‘Bay’ were sown in July 2013, and were grown in the field at Saga University under natural day light and standard fertilizer conditions (N = 0.6 kg/a; P = 0.6 kg/a; K = 0.6 kg/a). Green leaves and seeds were collected at several developmental stages, quickly frozen in liquid nitrogen, and stored at −80°C until DNA and RNA extraction. Mature seeds were also collected for analysis of fatty acid composition.
TILLING screeningWe screened 39,100 lines in five soybean mutant populations that corresponded to populations 1 to 5 (P1, X-ray irradiated population of ‘Bay’; P2, EMS treated population of ‘Bay’; P3, X-ray irradiated population of ‘Olerichi 50’; P4, X-ray irradiated population of ‘Fukuyutaka’; P5, EMS treated population of ‘FukuHOLL’) described in a previous review (Anai 2012). To identify non-functional alleles of GmFAD3-2a by high-throughput TILLING (Hoshino et al. 2010), we designed primers to target two amplicons covering the ORF of GmFAD3-2a (Fig. 1). In brief, the target nucleotide sequences were amplified with sequence-specific primer sets (5′ region: GmFAD3-2a-Full-F, 5′-GTGACATGTATGAATGATGTATCTTGTA-3′; GmFAD3-2a-INT1-R, 5′-GTGTGCGATGTATAAAGAAAGTGTGA-3′; 3′ region: GmFAD3-2a-Check-F1, 5′-TCCTTTCCCCATCTTTGCATACCCCT-3′; GmFAD3-2a-Full-R2, 5′-TCTAACATAGCATGCTTTGAAAAGTAAGTG-3′) (Fig. 1) from pooled template DNAs that consist of 5 to 8 independent mutant DNAs. After hetero-duplex formation through a heat-denature/annealing cycle, each amplicon was treated with a mismatch-specific nuclease, CEL I. Then, DNA fragments were prestained with the Gel Red dye (Wako) and separated by gel electrophoresis in 1.0 or 1.5% agarose with 1X TBE buffer. Each nucleotide sequence of PCR amplicon obtained from mutant candidate was determined with the Big Dye Terminator Cycle Sequencing Kit Ver. 3.1 (Applied Biosciences) using the same primers described above. Sequencing was performed by 3130 Genetic Analyzer (Applied Biosciences), and analyzed by Sequence Scanner software Ver. 1.0 (Applied Biosciences) and Genetyx-Mac Ver. 17.0.3 (Genetyx).

Allelic mutations in the GmFAD3-2a gene identified in soybean mutant lines by TILLING. Boxes represent exons (white, open reading frames; gray, untranslated regions). Nucleotide and amino acid substitutions between the wild-type cultivar ‘Bay’ (top) and mutants (bottom) are indicated. Numbers indicate the distance from the first nucleotide of the start codon or from the initial methionine. Arrows indicate the positions of gene-specific primers for PCR reactions.
Total RNA was isolated from leaves and seeds of ‘Bay’ and ‘P1-A9’ at each developmental stage in RNAiso Plus reagent (Takara Bio) according to the manufacturer’s instructions. First-strand cDNA was synthesized with a PrimeScript II first-strand cDNA synthesis kit (Takara Bio). The first-strand cDNA concentrations were normalized with the intensity of cDNA fragment amplified with an actin-specific primer set (SoyActin-F, 5′-GTTTGCGACAATGGAACAGGAATGGTTAAG-3′; SoyActin-R, 5′-TAATCTTCATGCTACTTGGGGC-3′) and separated in an agarose gel. The normalized cDNAs were used as templates for PCR to amplify the full-length region of GmFAD3-2a with sequence-specific primer sets (GmFAD3-2a-Full-F and GmFAD3-2a-Full-R2). PCR was performed as reported previously (Hoshino et al. 2011). The RT-PCR products were separated by gel electrophoresis in 1.5% agarose and visualized with EtBr staining.
Alignment of GmFAD3sThe deduced amino acid sequences of GmFAD3-1a, GmFAD3-1b, and GmFAD3-2a were obtained from GenBank (AB105886, AB105887, and AB051215, respectively; Anai et al. 2005). Alignment was generated with CRUSTALW (Thompson et al. 1994).
Yeast transformationTo evaluate the FAD3 enzyme activity of the novel GmFAD3-2a mutants, we expressed each of the wild-type (WT) and mutant alleles in budding yeast, and then analyzed the total fatty acid composition. Full-length cDNA fragments of GmFAD3-2a derived from ‘Bay’, ‘P1-A9’, ‘P3-B1’, and ‘P3-F4’ were amplified (GmFAD3-2a-Full-F and GmFAD3-2a-Full-R2) and cloned into the yeast expression vector pYES2/CT (Invitrogen) by a yeast-based recombination method (Nagano et al. 2007). The culture conditions for the recombinant yeast cells were described previously (Anai et al. 2005).
Fatty acid analysisYeast cells and soybean F3 seeds flour samples were directly methylated with acidic methanol and analyzed by gas chromatography as reported previously (Anai et al. 2008). Sample preparation was performed in triplicate and the statistical significance was calculated with the Tukey’s multiple comparison test.
Development of novel nonfunctional GmFAD3-2a alleles by using TILLING system is a valuable approach to reduce α-linolenic acid content in soybean seed oil. A total of 39,100 lines were applied to the CELI-based TILLING screening using two GmFAD3-2a-specific primer sets (Fig. 1). We identified seven independent mutations within the region of the GmFAD3-2a gene in the mutant lines and confirmed these mutations by direct sequencing of each PCR product (Fig. 1). Four lines contained silent mutations. Lines ‘P3-B1’, ‘P3-F4’, and ‘P1-A9’ contained missense mutations in exons of GmFAD3-2a: ‘P3-B1’ contained a single nucleotide change from guanine to adenine at nucleotide position 690 (g690a) in exon 5, which resulted in an amino acid substitution (Met230Ile). ‘P3-F4’ contained a mutation (c937t) in exon 7 with an amino acid substitution (His313Tyr). ‘P1-A9’ contained a mutation (c1047g) in exon 8 with an amino acid substitution (Ser349Arg).
Expression analysis of GmFAD3-2a in the mutantsSince mutations also occurred in the 5′-untranslated region (in ‘P3-B12’) or in introns (in ‘P3-D12’ and ‘P2-F1’) of GmFAD3-2a, they could affect gene expression or cause missplicing in these mutants. However, the expression patterns of GmFAD3-2a in leaves of ‘P3-B12’, ‘P3-D12’, and ‘P2-F1’ were similar to those of the WT (Fig. 2). The result suggests that any splicing variant would not be induced in these mutants. In contrast, ‘P1-A9’ contained an extra shorter-length cDNA fragment. Direct nucleotide sequencing revealed that the normal-size cDNA fragment contained a single nucleotide change (c1047g) resulting in the Ser349Arg substitution, and the shorter fragment contained a 95-bp deletion, also in exon 8 (Fig. 3). This deletion changed a 63-amino acid sequence into truncated 7-amino acid sequence in the COOH-terminal region (Fig. 4). This finding suggests that the truncated GmFAD3-2a protein of ‘P1-A9’ lacks enzyme activity, because the COOH-terminal region of FAD3 contains a signal for localization to the endoplasmic reticulum (ER) lumen (Dyer and Mullen 2001). These results strongly suggest that missplicing resulting from the same c1047g substitution of GmFAD3-2a in ‘P1-A9’ newly produced the shorter transcript. The presence of splicing variants of GmFAD3-2a in ‘P1-A9’ might affect its function relative to the ‘Bay’ allele. Therefore, we compared the amount of GmFAD3-2a transcript in leaves and seeds of ‘Bay’ and ‘P1-A9’ at several developmental stages. GmFAD3-2a transcripts were accumulated in both green leaves and developing seeds of these lines, and quantities decreased as the growth stage progressed (Fig. 5). The quantity of the normal-size transcript in the developing seeds was lower in ‘P1-A9’ than in ‘Bay’, although the transcript in young leaves of ‘P1-A9’ showed higher accumulation than that in ‘Bay’. This suggests that the missplicing due to the point mutation in ‘P1-A9’ affects the accumulation level of GmFAD3-2a transcripts in both young leaves and developing seeds. However, the enzyme activity of GmFAD3-2a product containing amino acid substitution in the ‘P1-A9’ was unknown.

Expression analysis of wild-type (WT) and GmFAD3-2a mutant soybean lines containing a mutation in an exon, 5′-untranslated region (UTR), or intron. Total RNA was prepared from green leaves and used in RT-PCR with gene-specific primers that targeted the full-length gene. Actin was used as the control.

Schematic diagram of spliced transcripts of GmFAD3-2a in wild-type (WT) and GmFAD3-2a mutant line ‘P1-A9’. Boxes represent exons (white, open reading frames; gray, untranslated regions). Nucleotide substitutions between the WT and ‘P1-A9’ are indicated (middle). Dashed white box in exon 8 (bottom) represents a deleted segment. SNP: single-nucleotide polymorphism.

Comparison of deduced amino acid sequences of three GmFAD3s from wild-type and GmFAD3-2a mutant line ‘P1-A9’. Asterisk indicates the amino acid residue mutated in ‘P1-A9’.

Tissue-specific expression of GmFAD3-2a in wild-type (WT) and GmFAD3-2a mutant line ‘P1-A9’. Total RNA was prepared from young leaves, mature leaves, and developing seeds at three different stages (<5 mm, 5–15 mm and >15 mm in diameter of developing seeds), and then was used for RT-PCR with gene-specific primer sets. These stages of developing seeds correspond to approximately 10, 20 and 30 days after flowering, respectively. Actin was used as the control.
To evaluate the functional consequence of the missense mutation causing the two variants of GmFAD3-2a in ‘P1-A9’, we compared the activity of microsomal FAD3 derived from ‘P1-A9’ and from ‘Bay’ by recombinant yeast system. Gas chromatography revealed no α-linolenic acid peak in yeast cells containing the empty vector (Fig. 6: pYES/CT). In contrast, an α-linolenic acid peak appeared at 8.6 min in yeast cells containing the WT allele of GmFAD3-2a (Fig. 6: WT). An identical peak appeared also in cells containing the ‘P3-B1’ and ‘P3-F4’ alleles. In contrast, cells carrying the ‘P1-A9’ allele and the ‘P1-A9Del’ splicing variant did not produce a detectable quantity of α-linolenic acid (Fig. 6: P1-A9 and P1-A9Del). These results clearly show that both ‘P1-A9’ mutant allele-derived transcripts could not produce functional FAD3 enzymes in yeast cells.
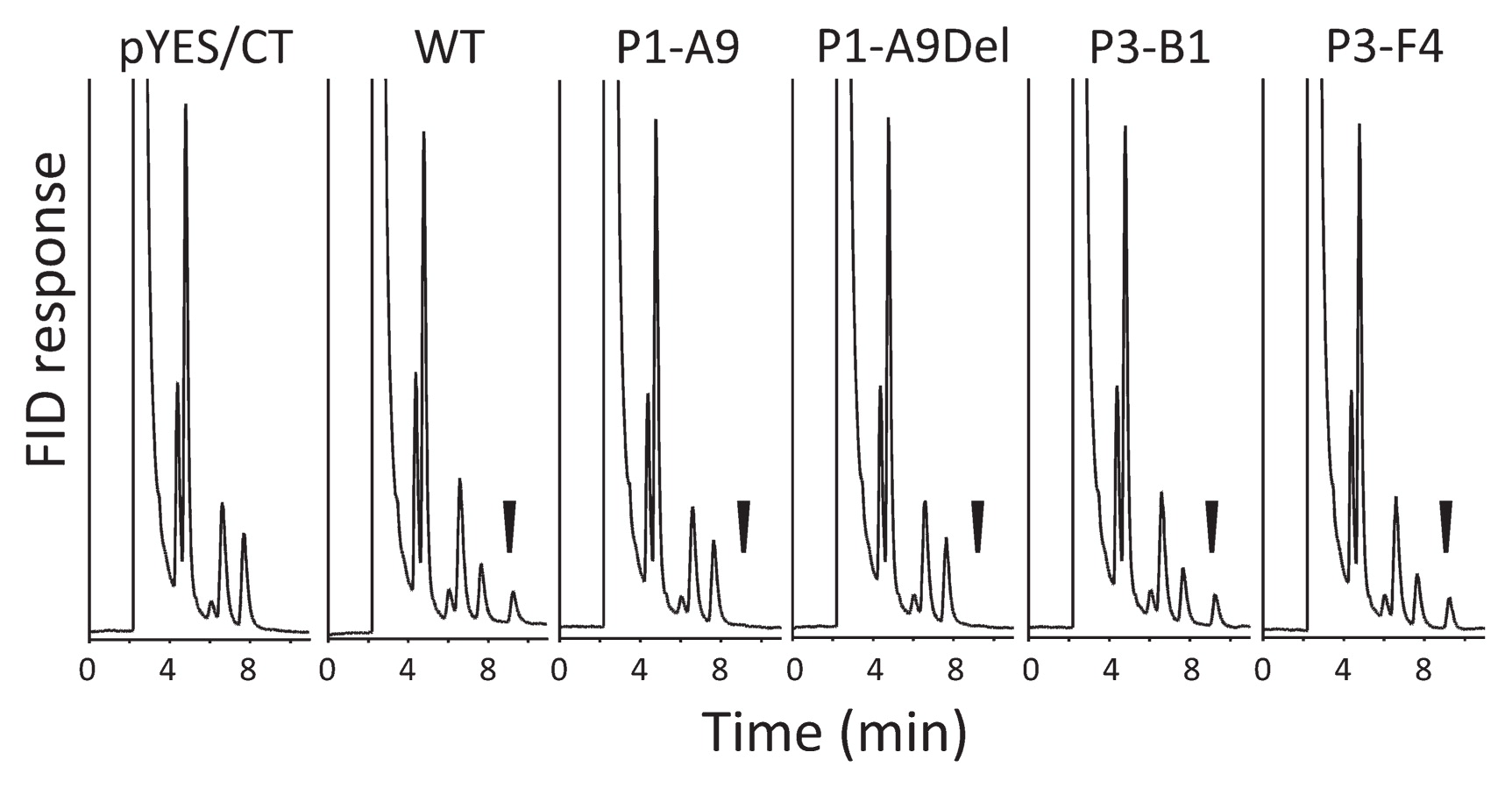
Functional analysis of wild-type (WT) and GmFAD3-2a mutant soybean genes expressed in yeast. Chromatograms of total fatty acids obtained from yeast cells harboring (pYES/CT) an empty vector or the same vector containing (WT) the ‘Bay’-derived GmFAD3-2a, (P1-A9) the mutant GmFAD3-2a containing a point mutation from ‘P1-A9’, (P1-A9Del) the mutant GmFAD3-2a containing a deletion in exon 8 from ‘P1-A9’, (P3-D1) the mutant GmFAD3-2a containing a point mutation from ‘P3-B1’, or (P3-F4) the mutant GmFAD3-2a containing a point mutation from ‘P3-F4’ by using flame ionization detector (FID). Arrowheads indicate α-linolenic acid.
To elucidate the contributions of the soybean FAD3 gene members to the biosynthesis of α-linolenic acid, we compared the fatty acid composition of the GmFAD3-2a mutant line ‘P1-A9’ with that of the GmFAD3-1a–GmFAD3-1b double-mutant line ‘LOLL’. In ‘Bay’, the α-linolenic acid content was 8.0% of the total fatty acids (Table 1). In ‘LOLL’, it was 3.1%, consistent with our previous result (Rahman et al. 1998). In ‘P1-A9’, it was 6.6%. However, in the triple-mutant ‘LOLL-A9’, it was <2%. These results well reflected the enzyme activities of the mutant products in yeast (Fig. 6), and indicate that GmFAD3-1a, GmFAD3-1b, and GmFAD3-2a together underlie FAD activity in soybean seed, as we predicted (Anai et al. 2005).
| Line | Palmitate | Stearate | Oleate | Linoleate | α-Linolenate |
|---|---|---|---|---|---|
| Bay | 12.8 ± 0.8 | 3.9 ± 0.1 | 22.4 ± 1.5 | 50.8 ± 1.4 | 9.4 ± 0.8** |
| P1-A9 | 12.8 ± 0.4 | 3.3 ± 0.4 | 24.4 ± 3.9 | 53.3 ± 4.1 | 6.2 ± 0.1** |
| LOLL | 14.4 ± 0.2 | 3.7 ± 0.5 | 22.7 ± 3.9 | 55.5 ± 3.9 | 3.6 ± 0.2* |
| LOLL-A9-1 | 14.1 ± 0.5 | 4.2 ± 0.6 | 22.8 ± 2.4 | 57.2 ± 2.3 | 1.7 ± 0.2 |
| LOLL-A9-2 | 14.3 ± 1.0 | 3.6 ± 0.4 | 25.3 ± 4.3 | 55.1 ± 3.2 | 1.7 ± 0.2 |
All plants were grown in the same field. Values are means ± SD obtained from three independent experiments.
Phenotype-based mutation screening of crops that have (paleo)polyploid genomes, such as soybean, is often limited by gene duplication, because the effect of a single-gene knockout is frequently masked by functional redundancy or environmental conditions. However, we have isolated valuable alleles of duplicated genes controlling oleic acid content (Hoshino et al. 2010) using our soybean TILLING system. Dierking and Bilyeu (2009) reported the isolation of new mutant alleles of a raffinose synthase gene and a microsomal omega-6 fatty acid desaturase gene through a similar system. The robustness and potential of this reverse genetics screening system has been validated by its successful application in many crop species, including wheat (Slade et al. 2005), rice (Suzuki et al. 2008, Till et al. 2007), barley (Caldwell et al. 2004), pea (Triques et al. 2007), and tomato (Minoia et al. 2010). No GmFAD3-2a mutant allele had hitherto been obtained from our X-ray-treated population by phenotype screening, likely because of the weaker activity of GmFAD3-2a than of the other FAD3 enzymes, and because the small phenotypic difference is easily masked by environmental factors. However, our successful isolation of the novel ‘P1-A9’ GmFAD3-2a allele by this reverse genetics approach shows that this robust system can obtain mutant alleles, depending on only their nucleotide polymorphism, and it is not affected by the degree of phenotypic change in the target gene product. Once we obtain several mutant candidate lines, it would become easy to evaluate relatively weaker phenotypes. Our results demonstrated that TILLING offers a nontransgenic alternative for the rapid generation of novel genetic variation. The application of TILLING to various target genes could help improve agricultural traits of soybean and other commercially important crops. Furthermore, this approach would provide a valuable and convenient resource in crops that have polyploidy genomes for not only commercial crop improvement but also basic functional genomic researches.
The GmFAD3-2a mutant allele that contains only one base substitution in ‘P1-A9’ generated two different transcripts (Figs. 2, 3). The major transcript encodes a protein similar to the WT, with an amino acid substitution (asterisk in Fig. 4) in a region highly conserved among the GmFAD3 family (Anai et al. 2005). This amino acid residue does not overlap with the ER-retention signal (Dyer and Mullen 2001), but we expected that the serine-to-arginine substitution would reduce or eliminate the enzyme activity of the GmFAD3-2a protein, because the polarities of these amino acids are markedly different. The results of GmFAD3-2a expression analysis in yeast clearly support our prediction (Fig. 6: P1-A9). On the other hand, the minor transcript encoded a frame-shifted product with a shorter COOH-terminal amino acid sequence. We speculate that this unusual splicing may be attributed to the partly altered recognition sites by splicing for the “AG” in front of the incidental base substitution in exon 8 as a 3′ splicing site. Because the COOH-terminal region of FAD3 contains a localization signal for the ER lumen (ER-retention signal) (Dyer and Mullen 2001), the translational product of this splicing variant might lack enzymatic activity owing to the loss of the ER-retention signal, as previously stated for ‘M5’ and ‘M24’ (Anai et al. 2005). Thus, we expected that neither GmFAD3-2a gene product of the ‘P1-A9’ allele would show any enzyme activity in planta.
GmFAD3-2a clearly contributed less to α-linolenic acid synthesis in soybean seeds than GmFAD3-1a and GmFAD3-1b, because α-linolenic acid contents were decreased to 82.5% of the control in ‘P1-A9’ (Table 1), 72.6% in ‘M24’ (GmFAD3-1a), and 53.2% in ‘M5’ (GmFAD3-1b) (Anai et al. 2005). In addition, because α-linolenic acid content is strongly affected by growth and environmental conditions (Dornbos and Mullen 1992, Wilcox et al. 1993), including flowering date and temperature, it might be difficult to isolate GmFAD3-2a mutants by phenotype screening.
As anticipated, the triple-mutant ‘LOLL-A9’ achieved <2% α-linolenic acid content (Table 1). Bilyeu et al. (2011) had already reported that the combination of three null mutant alleles—GmFAD3A (GmFAD3-1b), GmFAD3B (GmFAD3-1a), and GmFAD3C (GmFAD3-2a in this study)—generated ultra-low α-linolenic acid content. These mutant genes had been developed by induced mutagenesis and selected by phenotype screening (Bilyeu et al. 2005, Chappell and Bilyeu 2006). In our study, novel GmFAD3-2a mutant allele in ‘P1-A9’ was generated by TILLING, which can also provide useful genetic resources for developing elite soybean cultivars with ultra-low α-linolenic acid content from any desired cultivar.
Two of the most important goals in breeding soybeans for oil quality are to reduce the α-linolenic acid content and to increase the oleic acid content so as to achieve oxidative stability. Pham et al. (2012) reported to develop several soybean lines containing different mutant alleles of GmFAD3s and GmFAD2s, both of which encode microsomal omega-3 and omega-6 fatty acid desaturases, respectively, but no soybean line with all five mutant alleles has been reported yet. Because both α-linolenic acid and oleic acid contents are strongly influenced by growth temperature and modifier genes (Dornbos and Mullen 1992, Hyten et al. 2004, Wilcox et al. 1993), pyramiding of GmFAD2 and GmFAD3 mutant alleles will lead to improvement in soybean oil quality.
In this study, we have demonstrated the ability of TILLING to isolate a weak functionally overlapping gene controlling α-linolenic acid content of soybean seeds. This result shows the capacity of a mutant-based reverse genetics strategy to develop novel minor but effective alleles for improving various crop traits.
This work was partially supported by a Grant-in Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan to TA.