2015 Volume 65 Issue 5 Pages 438-446
2015 Volume 65 Issue 5 Pages 438-446
Indonesia is the third largest cocoa-producing country in the world. Knowledge of genetic diversity and parentage of farmer selections is important for effective selection and rational deployment of superior cacao clones in farmers’ fields. We assessed genetic diversity and parentage of 53 farmer selections of cacao in Sulawesi, Indonesia, using 152 international clones as references. Cluster analysis, based on 15 microsatellite markers, showed that these Sulawesi farmer selections are mainly comprised of hybrids derived from Trinitario and two Upper Amazon Forastero groups. Bayesian assignment and likelihood-based parentage analysis further demonstrated that only a small number of germplasm groups, dominantly Trinitario and Parinari, contributed to these farmer selections, in spite of diverse parental clones having been used in the breeding program and seed gardens in Indonesia since the 1950s. The narrow parentage predicts a less durable host resistance to cacao diseases. Limited access of the farmers to diverse planting materials or the strong preference for large pods and large bean size by local farmers, may have affected the selection outcome. Diverse sources of resistance, harbored in different cacao germplasm groups, need to be effectively incorporated to broaden the on-farm diversity and ensure sustainable cacao production in Sulawesi.
Cacao (Theobroma cacao L.), the source of cocoa butter and powder for the confectionery industry, is an important tropical crop. Together with major quantities of milk, sugar, almonds and peanuts, it supports a multi-billion dollar chocolate industry. Worldwide, the product obtained from dried fermented cacao seeds (i.e. cocoa) is a $5 to $6 billion export commodity, of which 90% is produced by smallholder farmers in tropical developing countries (World Cocoa Foundation 2012). Indonesia is the third largest cocoa-producing country after Ivory Coast. With an annual production of 740,500 metric tons of cocoa beans, it constitutes about 16% of world cocoa bean production (FAOSTAT 2012, production data available at http://faostat3.fao.org/faostat-gateway/go/to/browse/Q/*/E). Indonesia’s cocoa exports are valued at approximately $1.2 billion per year and provide the main source of income and livelihood for over 400,000 smallholder farmers, with farm sizes ranging from 0.5–2 hectares (Agriculture Department of Indonesia 2008). Among the cocoa producing regions in Indonesia, the island of Sulawesi is the largest, accounting for 71% of the annual output of the entire nation or nearly 13% of the world’s supply. In the last 20 years, cocoa production in Sulawesi has experienced rapid increase, largely based on the expansion of cultivation area. However, the average yield has been stagnant at the level of about 700 kilos per hectare (Agriculture Department of Indonesia 2008). Lack of improved varieties and planting materials is one of the main production constraints (Neilson 2007, Perdewa and Shively 2009, Ruf and Yoddang 2010). Traditionally, planting materials used in Sulawesi were mostly hybrids and clone propagation has been rare until recent years (ACDI/VOCA 2005). The sources vary, and include hybrids produced in seed gardens operated by Indonesian government, local selections made by Sulawesi farmers from their own farms, and materials introduced by migrant workers from plantations in Malaysia (Iswanto et al. 1997). As an out-crossing species, hybrid cacao seedlings show segregation in phenotypic and agronomic traits (Dias 2001, Silva et al. 2011). Usually, only a fraction of the cacao trees in a farm produces decent yield, and overall yield and quality of cacao beans are affected. Other important production constraints include diseases and pests such as vascular streak dieback (VSD, Oncobasidium theobromae), black pod rot (Phytophthora palmivora) and cocoa pod borer (Conopomorpha cramerella) (Keane 1992, McMahon et al. 2009, Wardojo 1992). Genotypes resistant to VSD and black pod rot have been identified, but delivery of the appropriate genetic material to farmers’ fields remains a major challenge (McMahon et al. 2009, Susilo 2009).
Recent programs to improve cocoa production through rehabilitation and intensification have brought a new incentive to use high-yielding and disease-resistant clones in Sulawesi (Ruf and Yoddang 2010, Susilo 2009). Replacing low-yielding cacao trees with superior clones is an effective approach to boost yield for small holders (ACDI/VOCA 2005, McMahon et al. 2009). Supported by the Indonesian government, the chocolate industry and various organizations in the international community, superior cacao clones from farmers’ fields and government breeding programs have been evaluated, propagated, and adopted by farmers, often through side-grafting and chupon-grafting (Susilo et al. 2009). With the subsidy from the Indonesian government, mass production of superior clones through embryogenesis has produced impressive results (Arifin 2013, Cocoa Sustainability Partnership 2013). Meanwhile, local nurseries are also propagating farmer selections identified from local farms and communities, playing a complementary role to the centralized seeds system (Susilo et al. 2009).
Indonesia used to grow Trinitario type cacao before the 1950s (Bartley 2005, Toxopeus and Geisberger 1983). However, the Trinitario cacao lacked resistance to cacao diseases and pests, especially to VSD, which led to the breeding of bulk-cocoa starting in the 1950s, using parental clones from Upper Amazon Forastero (Iswanto et al. 1997, Toxopeus and Geisberger 1983). In the mid 1970s more hybrids of Upper Amazon Forastero were introduced from Malaysia and they were widely adopted by growers in North Sumatra (Mawardi et al. 1994, Toxopeus and Geisberger 1983). With the enrichment of these introduced materials, new germplasm with tolerance to black pod and VSD were developed and used either in government supported seed gardens or released as new varieties, which supported the rapid expansion of cacao production in Indonesia over several decades (Iswanto et al. 1997, McMahon et al. 2009, Susilo et al. 2009). Farmer participatory selection has been advocated as an efficient approach to complement centralized breeding programs in various crops (Bellon 2004). Participatory selection may be more useful for tropical perennial crops in low input and small scale production systems. Farmers’ experience in multi-year field observations can identify superior trees, because specific adaptation receives adequate weight as a criterion for selection (Dias 2001, Eskes 2006). To use this breeding scheme, the underlining assumption is that high levels of genetic diversity and variability in agronomic traits exist in farmers’ fields, which can be explored for the identification of promising clones (Dias 2001). Knowledge about the genetic background of farmer selections is therefore important for effective selection and rational deployment of superior clones. Moreover, quality control for clonal propagation is essential to ensure the accuracy and reliability of the planting materials. The objective of the present study was to assess the genetic background of local farmer selections using SSR markers in order to understand their ancestry and genetic variability. The resultant information will help selection of superior trees from farmers’ fields and is essential for the on-going program of rehabilitation of old cacao farms in Sulawesi, Indonesia.
A total of 53 local farmer selections together with 152 international clones were used in this experiment (Table 1). The farmer selections were collected by agricultural research institutes and private companies of chocolate industry, and farmer communities in Sulawesi. The genetic basis of these farmer selections was mainly hybrid families from various farmer seed gardens. The parental clones in these seed gardens include breeding lines developed by Indonesian Coffee and Cocoa Research Institute (ICCRI), introduced clones brought in from Papua New Guinea and Malaysia, and local Trinitario clones that have survived despite the heavy diseases (especially VSD) in Sulawesi (Keane and Prior 1991). This on-farm diversity provides an opportunity for farmer participatory selection and propagation of superior clones. Beginning in the late 1990s, a number of high-yielding trees were selected from farmers’ fields (Mawardi et al. 1994, Susilo et al. 2009). These local selections were then side-grafted onto mature trees in the germplasm nurseries in Indonesian Coffee and Cocoa Research Institute (ICCRI) in Java and in Mars Cocoa Development Center (MCDC) in Makassar (McMahon et al. 2009, Susilo et al. 2009). All the farmer selections used in the present study were considered by the providers as promising clones, either for direct deployment or for breeding, and a fraction of them are currently being evaluated in multi-location field trials.
| # | Clones | Source |
|---|---|---|
| 1 | ACC | MCDCa |
| 2 | AM | MCDC |
| 3 | AP | MCDC |
| 4 | BAMBU | MCDC |
| 5 | BB | MCDC |
| 6 | BRT | MCDC |
| 7 | FQ | MCDC |
| 8 | GENIJ | MCDC |
| 9 | ISMAIL | MCDC |
| 10 | KAMBALA | MCDC |
| 11 | KW516 | MCDC |
| 12 | KW617 (a) | MCDC |
| 13 | KW617 (b) | MCDC |
| 14 | L1 | MCDC |
| 15 | LG | MCDC |
| 16 | M 01 | MCDC |
| 17 | M 02 | MCDC |
| 18 | M 04 | MCDC |
| 19 | M 05 | MCDC |
| 20 | M 06 | MCDC |
| 21 | M 07(a) | MCDC |
| 22 | M 07(b) | MCDC |
| 23 | M 08 | MCDC |
| 24 | MY03 | MCDC |
| 25 | PR | MCDC |
| 26 | PWPQ | MCDC |
| 27 | RB(a) | MCDC |
| 28 | Sulawesi 1 | MCDC |
| 29 | Sulawesi 2 | MCDC |
| 30 | BTR | ICCRIb |
| 31 | PW/PG | ICCRI |
| 32 | HARIJ | ICCRI |
| 33 | Hasbi Tari | ICCRI |
| 34 | ILH | ICCRI |
| 35 | KDI1 | ICCRI |
| 36 | KDI2 | ICCRI |
| 37 | RB (b) | ICCRI |
| 38 | MT | ICCRI |
| 39 | KW (unknown) | ICCRI |
| 40 | Moktar | ICCRI |
| 41 | Patila | ICCRI |
| 42 | Toli-Toli | ICCRI |
| 43 | YD 75 | ICCRI |
| 44 | YM | ICCRI |
| 45 | ARDACIAR 10 | ICCRI |
| 46 | ARDACIAR 26 | ICCRI |
| 47 | Nasir Rauf | ICCRI |
| 48 | Panimbu Red | ICCRI |
| 49 | PCK | ICCRI |
| 50 | Pengawu | ICCRI |
| 51 | Sausu Piore | ICCRI |
| 52 | SS Rewang | ICCRI |
| 53 | Terobok Green | ICCRI |
| Reference cacao clones (with known genetic identities) used in data analysis | |||
|---|---|---|---|
| # | Germplasm groups | No. of samples | Served as reference in data analysis |
| 1 | IMC | 23 | Clustering, Bayesian assignment and parentage analysis |
| 2 | Nanay | 22 | Clustering, Bayesian assignment and parentage analysis |
| 3 | Parinari | 22 | Clustering, Bayesian assignment and parentage analysis |
| 4 | Morona | 14 | Clustering, Bayesian assignment and parentage analysis |
| 5 | SCA & Ucayali | 14 | Clustering, Bayesian assignment and parentage analysis |
| 6 | Nacional hybrids | 12 | Clustering analysis and Bayesian assignment |
| 7 | Trinitario | 21 | Clustering analysis only |
| 8 | Amelonado | 16 | Clustering analysis only |
| 9 | Criollo | 8 | Clustering analysis only |
| Total reference | 152 | ||
Leaves collected from individual cacao trees on local farms were used for DNA fingerprinting profiles. Two healthy young leaves were collected from each tree, and the samples were dried in silica gel and sent to the USDA Beltsville Agricultural Research Center, Maryland, USA for genotyping. Cacao DNA was extracted from the leaf tissue using the DNeasy Plant System (Qiagen Inc., Valencia, CA, USA) according to Saunders et al. (2004). For the 152 international clones, the SSR data were generated in previous experiments. These international clones were included in this experiment as reference clones. The genetic identities of these clones were determined through an international initiative of DNA fingerprinting of cacao (Susilo et al. 2011, Zhang et al. 2009a, 2009b). The majority of the reference clones were maintained in the two international genebanks in Trinidad and Costa Rica (Motilal et al. 2010, Zhang et al. 2009a, 2009b). The rest of the reference clones were from the cacao germplasm collection in the Indonesian Coffee and Cocoa Research Institute (ICCRI), Agricultural Research Institute (INIAP) of Ecuador and Tropical Crop Institute of Peru (ICT) of Peru.
SSR markers and genotypingAmplification of microsatellite loci was achieved using 15 primers with sequences previously described (Lanaud et al. 1999, Risterucci et al. 2000, Saunders et al. 2004). These 15 loci have been agreed upon by multiple international and government-sponsored laboratories in the cacao research community, as standardized SSR primers to characterize all T. cacao germplasm collections (Saunders et al. 2004). These standard loci have been used for cacao genotyping in several germplasm collections (Zhang et al. 2006, 2009a, 2009b). Primers were synthesized by Proligo (Boulder, CO) and forward primers were 5′-labeled using WellRED fluorescent dyes (Beckman Coulter, Inc., Fullerton, CA). PCR was performed as described in Saunders et al. (2004), using commercial hot-start PCR SuperMix that had been fortified with an additional 30 U/ml of hot-start Taq DNA polymerase (Invitrogen Platinum Taq, Carlsbad, CA, or Eppendorf HotMaster Taq, Brinkman, Westbury, NY).
The amplified microsatellite loci were separated by capillary electrophoresis as previously described (Saunders et al. 2004, Zhang et al. 2006). Data analysis was performed using the CEQ™ 8000 Fragment Analysis software version 7.0.55, according to manufacturers’ recommendations (Beckman Coulter, Inc.). SSR fragment sizes were automatically calculated to two decimal places by the CEQ™ 8000 Genetic Analysis System. Allele calling was performed using the CEQ™ 8000 binning wizard function, version 7.0.55 (Beckman Coulter, Inc.) and, based on the bin list, edited using SAS software (SAS 1999). Two of the 152 reference clones were re-run with the Sulawesi farmer selections in order to provide an internal check for consistency of SSR fragment sizing.
Analysis of genetic diversity and parentage background using SSR dataSummary descriptive statistics were computed for this group of farmer selections. The descriptive statistics included the number of alleles, observed and expected heterozygosity and inbreeding coefficient. All statistics were calculated using GenAlEx 6.2 (Peakall and Smouse 2006, Peakall and Smouse 2012).
Cluster analysis was used to examine the genetic relationship among the same clones. Kinship coefficient among individual clones was calculated using the program Microsatellite Analyser (Dieringer and Schlötterer 2003). A dendrogram was generated from the resulting distance matrix using the Neighbor-Joining algorithm (Saitou and Nei 1987) available in PHYLIP (Felsenstein 1989), and visualized using the program FigTree version 1.4 (Rambaut 2009).
For the analysis of population structure and inference of admixed ancestry, we used a model-based clustering method implemented in the software program STRUCTURE (Pritchard et al. 2000). Seven reference populations, representing all known genetic groups presented in Indonesia, were used in the analysis to trace the possible ancestry in these farmer selections. The germplasm groups of Criollo and Amelonado were excluded in the ancestry inference, because these two groups were not used as parents in Indonesia’s breeding program and seed gardens. The traditional variety “Java Criollo” is actually a Trinitario-type variety thus was included in the Trinitario reference group.
The K value was set from 1 to 10 and the analysis was carried out without assuming any prior information about the genetic group or geographic origin of the samples. We used STRUCTURE with 200,000 iterations after a burn-in period of 100,000. Ten independent runs were assessed for each fixed number of clusters (K). The ΔK value was computed to detect the most probable number of clusters (Evanno et al. 2005). Of the 10 independent runs, the one with the highest Ln Pr (X|K) value (log probability or log likelihood) was chosen and represented as bar plots.
To further illustrate the parentage contribution of the introduced germplasm since 1950s, parentage analysis was applied as a complementary method using SSR data. For this analysis the 53 farmer selections were used as offspring, whereas the reference clones were wild cacao from IMC, NA, PA, MO and SCA (n = 95, Supplemental Table 1). These 95 wild cacao clones included all the known progenitors in seeds gardens or breeding programs in Indonesia since the 1950s. Thus the analysis will allow us to assess the parentage contribution from the wild Upper Amazon Forastero, which is known to carry some disease resistances. A likelihood-based method, implemented in the program CERVUS 3.0 (Kalinowski et al. 2007, Marshall et al. 1998), was used for computation. For each parent-offspring pair, the natural logarithm of the likelihood ratio (LOD score) was calculated. Critical LOD scores were determined for the assignment of parentage to a group of individuals without knowing the maternity or paternity. Simulations were run for 10000 cycles. Assuming that 60% of candidate parents were sampled, the analysis typed 90% of the loci. The two most probable mother (or father) possibilities for each offspring were identified on the basis of the critical difference in LOD scores (Δ) between the most likely and next most likely candidate parent, at greater than 95% or 80% confidence (Kalinowski et al. 2007, Marshall et al. 1998).
The descriptive statistics of genetic diversity is presented in Table 2. The level of allelic diversity in the farmer selections was moderate. The average number of alleles in the Sulawesi samples was 8.67 whereas the reference group had 11.60. Moreover, the reference group had 4.2 private alleles, whereas none were observed in the Sulawesi group. The Sulawesi group had higher observed heterozygosity (Ho = 0.580) and lower inbreeding coefficient (F = 0.131) than the reference group (Ho = 0.419, F = 0.283) indicating the hybrid feature of the Sulaswesi farmer selections. Pairwise comparison of all individual clones showed a high level of genotype diversity among the farmer selections. The 53 farmer selections are all distinctive clones without duplication.
| Pop | No. of alleles | Observed Heterozygosity | Expected Heterozygosity | Inbreed. coefficient |
|---|---|---|---|---|
| Sulawesi farmer selections (n = 53) | ||||
| Mean | 8.67 | 0.580 | 0.667 | 0.131 |
| SE | 0.88 | 0.039 | 0.038 | 0.034 |
| Reference clones (n = 152) | ||||
| Mean | 11.6 | 0.419 | 0.740 | 0.283 |
| SE | 0.96 | 0.024 | 0.030 | 0.018 |
The neighbor-joining tree showed that the 205 cacao clones (53 Sulawesi farmer selections and 152 reference clones) were separated into two main clusters. The first cluster consisted of reference clones only, including wild cacao germplasm collected in the Peruvian Amazon (Zhang et al. 2009b), i.e Scavina, Morona, IMC and the Nacional hybrids from Ecuador. The second cluster included all the Sulawesi farmer selections, three traditional varieties (Trinitario, Amelonado and Criollo) and two wild cacao germplasm groups (Nanay and Parinari) (Fig. 1). Among them, Criollo, Amelonado and Trinitario have been existing in Indonesia for more than 400 years (Bartley 2005, Susilo et al. 2009), whereas the Nanay and Parinari groups were introduced in the 1950s.

Neighbor-joining dendrogram depicting the relationship between 53 farmer selections from Sulawesi, Indonesia, and 152 reference clones. Kinship coefficient was used as genetic distances. All Sulawesi clones correspond to the sample list in Table 1. All reference international clones correspond to the sample list in Supplemental Table 1.
Based on the result of ΔK (Fig. 2, Evanno et al. 2005), Bayesian clustering analysis led to the grouping of the reference clones into five most probable clusters, including Trinitario, IMC, Nanay, MO & SCA and Parinari (Fig. 3). The five clusters represented all known germplasm groups as possible founders for the current on-farm diversity in Sulawesi. The reference population of Refractario (Nacional hybrids) was shown as hybrids between Morona genotypes and Trinitario. On average, the five reference clusters have a coefficient of membership (Q value) of 0.926. A Q value of 0 corresponds to an individual of purely exogenous origin, whereas a value of 1 signifies the individual is purely from a home cluster. The majority of the 53 farmer selections have a hybrid genotype, with their admixed ancestry coming from two to four of the five reference germplasm groups (Fig. 3). Among the 53 farmer selections, only six genotypes can be classified as single ancestral origin (Q value ≥ 0.75), and all six were Trinitario (Ismail, PCK, Moktar, KDI 1, YD 75 and ARD ACARI 26). The remaining 47 selections were classified as interpopulation hybrids, which showed combinations of different ancestries (Fig. 3).
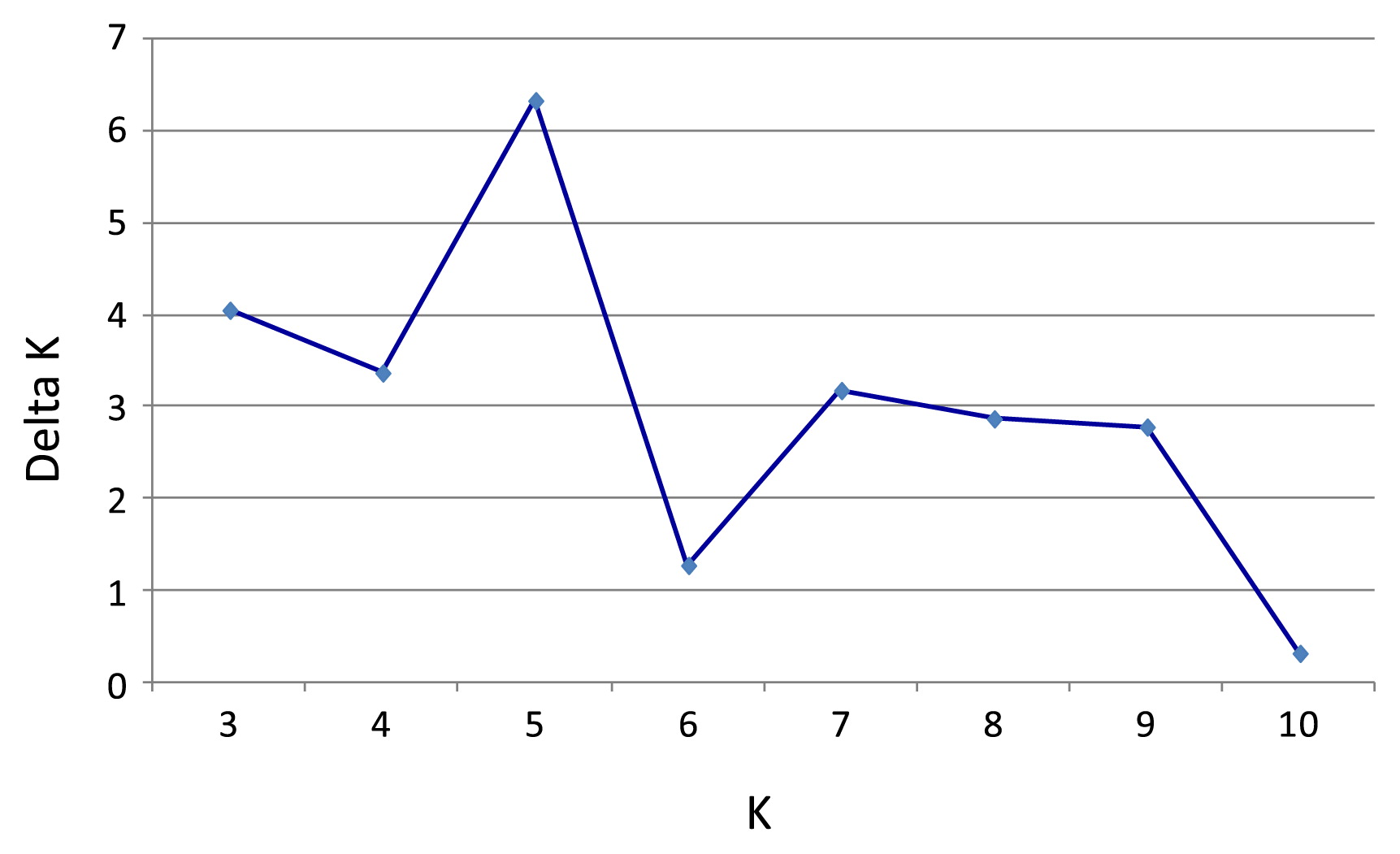
Plot of Delta K (filled circles, solid line) calculated as the mean of the second-order rate of change in likelihood of K divided by the standard deviation of the likelihood of K, m(|L00(K)|)/s [L(K)].

Inferred clusters in the Sulawesi farmer selections and reference clones using STRUCTURE, where K is the potential number of genetic clusters that may exist in the overall sample of individuals. Each vertical line represents one individual multilocus genotype. Individuals with multiple colors have admixed genotypes from multiple clusters. Each color represents the most likely ancestry of the cluster from which the genotype or partial genotype was derived. Clusters of individuals are represented by colors.
Among the five germplasm groups, Trinitario made the largest ancestral contribution to these farmer selections, with an average population membership (Q value) of 51.0% (Fig. 4). The second largest contributor was from the Parinari group, which explained 27.5% of the ancestry. The Nanay group accounted for 12.5% of the admixture, and the group of Morona/Scavina and IMC only explained 4.3% and 4.7% of the assigned membership of the 53 farmer selections (Fig. 4). This result is largely compatible with the previous result of multivariate analysis and further demonstrates that a majority of the Sulawesi farmer selections are hybrids of Trinitario and two Upper Amazon Forastero progenitors, especially those from the Parinari group. The parental contribution from Morona/Scavina was negligible.

Partitioned ancestry based on average memberships (Q-Value) of assigned founder germplasm groups in the Sulawesi farmer selections.
Results of the likelihood-based parentage analysis was presented in Table 3. For the 53 farmer selections, nine parent-offspring relationships, contributed by six parental clones, were identified at the confidence level above 80%. When the confidence level was raised to 95%, the number of parent-offspring relationships was reduced to six (Table 3). Among the nine identified parent-offspring relationships, five were associated the germplasm group of Nanay and four with Parinari. The parental clone NA 32 was found responsible for four offspring, including clone “Sulawesi 1” (original name “PBC 123”), one of the most popular clones in Southeast Asia, as well as for “Sulawesi 2” (original name “BR 25”), another popular clone in Sulawesi. No parental clone from the Scavina and Morona group was identified. The result of parentage analysis fully agreed with the results of NJ-tree and Bayesian test in terms of the assigned membership of the founder populations or germplasm groups.
| Offspring ID | Identified candidate mother/father | LOD scorea |
|---|---|---|
| M 07 | NA 226 | 8.96 |
| Pengawu | NA 32 | 7.08 |
| Sulawesi 2 | NA 32 | 10.00 |
| Sausu Piore | NA 32 | 9.87 |
| Sulawesi 1 | NA 32 | 9.30 |
| ARDACIAR 10 | PA 7 | 4.50 |
| KDI 2 | PA 300 | 21.12 |
| PWPQ | PA 303 | 4.38 |
| PR | PA 71 | 4.97 |
The high level of genotype diversity (i.e. each selection has a unique genotype) and observed heterozygosity in the 53 farmer selections indicated that these selections were derived from hybrid families, which was consistent with the history of germplasm introduction in Sulawesi (Iswanto et al. 1997, Toxopeus and Geisberger 1983). Cacao planting materials in Sulawesi were traditionally distributed by pods (Bartley 2005). Cacao propagation by clones was not common until recently. As an out crossing species, the segregating hybrids families offer a high level of heterozygosity and low inbreeding coefficient in the cacao fields. Nonetheless, the allele richness (8.67 alleles per locus) in the 53 farmer selections was moderate, and lower than the reference cacao clones representing seven different genetic clusters (11.60 alleles per locus in 152 clones). The expected heterozygosity was slightly lower in the farmer selections, relative to the reference clones. There was a larger difference between the observed heterozygosity and expected heterozygosity in the farmer selections. This difference was expected because most of the reference clones were from wild populations, whereas the farmer selections had likely gone through assorted mating and selections.
Results of the kinship based NJ tree and Bayesian clustering analysis of genetic diversity and ancestry showed that the farmer selections from Sulawesi are mainly hybrids from Trinitario and Upper Amazon Forastero. The result is in accordance with Indonesia’s cacao breeding history. In Indonesia, breeding of bulk-cocoa varieties using parental clones of Upper Amazon Forastero started in the 1970s, due to the demand for resistance to vascular streak die back disease (Iswanto et al. 1997, Toxopeus and Giesberger 1983). In the early 1950s, several Upper Amazonian clones from the genetic groups of Scavina, Parinari, and Nanay were introduced and a number of these clones were used in Indonesia’s breeding programs in the mid-1950s. In addition, there has been significant introduction of improved varieties from Malaysia. It is well known that the cacao varieties were often introduced from Malaysia by the Bugis migrant farmers in Sulawesi (Durand 1995). These farmers gained knowledge of the new hybrid cacao and of the farming techniques in Sabah, Malaysia, where Upper Amazonian Forastero and Trinitario clones were commonly used parental clones (Durand 1995).
However, the overall scope of genetic diversity in these farmer selections accounts for only a small fraction of the available germplasm groups in the reference clones. The revealed parentage background was mainly limited to a few clones in the Parinari germplasm group, and to a less dirversity in the Nanay group. The other groups, such as SCA and MO had almost no impact. The SCA clones are wild cacao collected from the Peruvian Amazon and have served as the major source of disease resistance in modern cacao breeding, including for witches’ broom disease (Moniliophthora perniciosa), black pod rot (Phytophthora spp.) and frosty pod (Moniliophthora roreri). According to the International Cacao Germplasm Database, “SCA 6” has been used by more than 14 breeding programs around the world. SCA 6 and SCA 12 have been consistently used in breeding program and seeds gardens in Indonesia as well, for their resistance to VSD and black pod (Susilo et al. 2009). In recent years, SCA 6 has been promoted in Sulawesi through embryogenesis. In spite of the common use of parental clones from MO/SCA germplasm groups in breeding and seeds gardens, these germplasm groups were not well incorporated into the farmer selections. This disparity could be due to limited access of the farmers to diverse planting materials. It’s also possible that the strong preference for large pods and large bean size by local farmers may have affected the selection outcome. The farm gate price for large beans is about 10 cents per kilo higher than for small beans in Indonesia. This preference for the exterior attractiveness of the pods and beans, with less attention on disease resistance, may undermine the efforts to incorporate diverse disease resistance into the cacao plantings in Sulawesi. From the perspective of breeding, recurrent selection would help the introgression of disease resistance but still retain the attractive pod and bean size.
In summary, microsatellite markers were used to assess genetic identity and parentage background of 53 farmer selections from Sulawesi. We demonstrated the Sulawesi farmer selections are mainly comprised of hybrids derived from three cacao germplasm groups: Trinitario and two Upper Amazon Forastero groups. This genetic foundation accounts for only a small fraction of available cacao genetic diversity, implying a narrow range of host resistance to cacao diseases and pests. New sources of resistance from other germplasm groups need to be introduced to broaden diversity in cacao germplasm and ensure sustainable cocoa bean production in Sulawesi. The present study provides important baseline information for selecting the most promising clones from farmers’ fields and ensuring genotype conformity in clone propagation. These are both essential for the on-going program of cacao rehabilitation and intensification in Sulawesi.
The authors would like to thank Stephen Pinney and Yan Mei-Li of USDA-ARS, SPCL for assistance in DNA sample preparation and SSR genotyping, and Virginia Sopyla of the World Cocoa Foundation for logistical support. The authors would also like to acknowledge USDA-FAS and the World Cocoa Foundation for the Borlaug Cocoa Fellowship program.
Special thanks are due to Pak Ismail, Sahardi Mulia, Hussin bin Purung and Smilja Lambert for their assistance in collecting leaf samples and providing background information of the local farmer selections in Sulawesi. Two anonymous reviewers are thanked for their review of the manuscript and suggestions for revision.