2016 Volume 66 Issue 5 Pages 776-789
2016 Volume 66 Issue 5 Pages 776-789
A pair of complementary genes, Hwc1-1 at HWC1 locus and Hwc2-1 at HWC2 locus, cause a weakness phenomenon in rice. For this study, we performed haplotype analysis around the HWC2 locus in two core collections comprising 119 accessions. We also examined reactions to phenol and Xanthomonas oryzae pv. oryzae (Xoo) Japanese race I. To elucidate the genetic relations among all accessions, we analyzed their banding patterns of 40 Indel markers covering the rice genome. The classification by Indel markers was almost consistent with that using 4,357 SNPs. The testcross with Hwc1-1 carrier indicated that 37 accessions carried Hwc2-1 allele, whereas 82 carried hwc2-2 allele. Strong association between HWC2 and Ph genes was observed. Based on 14 DNA markers around HWC2 locus and Ph genotype, the 119 accessions were divided into 50 haplotypes. To examine the HWC2 candidate chromosomal region specifically, the ‘haplotype group’ characterized by the six DNA markers closely linked with HWC2 were analyzed. Hwc2-1 carriers had the same haplotype group. Some hwc2-2 haplotype groups were associated with resistance against the Xoo race. The relation between varietal differentiation and haplotypes around the HWC2 locus was discussed, along with its breeding significance.
Asian cultivated rice (Oryza sativa L.) has often been classified into two varietal groups: Indica and Japonica. The Japonica group is classified further into temperate Japonica and tropical Japonica. This classification has been made according to morphological and physiological characters (Oka 1953, Sato 1991). Electrophoretic banding patterns of isozymes classify rice cultivars into two major groups corresponding to Indica and Japonica (Glaszmann 1987, Ishikawa et al. 1991). DNA marker techniques such as RFLP, SSR, Indel, and SNP also classify rice cultivars into two major groups corresponding to Indica and Japonica (Doi et al. 2000, Ebana et al. 2010, Garris et al. 2005, Huang et al. 2012, Lu et al. 2009, Xiong et al. 2010, Xu et al. 2012, Zhao et al. 2010, 2011). Some studies have divided Indica and Japonica into several subgroups. For example, using 169 SSR markers and 234 cultivars, Garris et al. (2005) classified Japonica into temperate japonica, tropical japonica, and aromatic, and classified Indica into aus, which corresponds to a minor group designated as II by Graszmann (1987), and indica. For the present study, to avoid confusion, ‘Indica’ is assigned to one of two major varietal groups, the other of which is Japonica, whereas ‘indica’ is assigned to one of subgroup in Indica.
Weak growth occurring in hybrids derived from crosses between two normal accessions is called hybrid weakness. According to its degree or symptom, it is also called hybrid lethality, hybrid abnormality, and hybrid necrosis. This phenomenon has been observed in many plant species including Arabidopsis thaliana (Bomblies et al. 2007) and Hordeum vulgare (Konishi 1985) (for a review, Bomblies and Weigel 2007). In many cases, it is controlled by a set of non-allelic complementary dominant genes. Amemiya and Akemine (1963) found a hybrid weakness caused by a set of dominant complementary genes in rice, Hwc1-1 at the HWC1 (HYBRID WEAKNESS C1) locus and Hwc2-1 at the HWC2 (HYBRID WEAKNESS C2) locus. Skewed distribution of Hwc2-1 genes toward temperate Japonica cultivars has been reported (Sato and Hayashi 1983, Sato and Morishima 1987). Among the examined cultivars, most temperate Japonica cultivars carry Hwc2-1, although few tropical Japonica and Indica cultivars carry this gene (Sato and Hayashi 1983, Sato and Morishima 1987). None of the 30 accessions of wild relatives (O. rufipogon and O. nivara) carries Hwc2-1 (Sato and Morishima 1987). Regarding HWC1, a tropical Japonica cultivar from Peru, ‘Jamaica’, is reportedly the only carrier of Hwc1-1 gene (Sato and Morishima 1987). From these results, Sato and Morishima (1988) inferred that the Hwc2-1 gene arose at an early stage of differentiation of temperate Japonica cultivars. A skewed distribution of Hwc2-1 genes toward temperate Japonica cultivars suggests the presence of a linkage block distinguishing temperate Japonica from other varietal groups around the HWC2 locus.
We mapped HWC2 on chromosome 4 between the two DNA markers, KGC4M28 and RM5473, and showed that the two genes, Ph (Phenol staining) and XA1 (XANTHOMONAS ORYZAE PV. ORYZAE RESISTANCE 1), are linked closely with HWC2 (Ichitani et al. 2001, Kuboyama et al. 2009) (Fig. 1). Phenol reaction is a useful criterion for classifying rice cultivars into Indica and Japonica (Oka 1953, Sato 1991): most Indica cultivars carry dominant alleles Ph: their grains are stained black, reacting to phenol. Most Japonica cultivars carry recessive alleles ph: their grains remain their original color. According to Oryzabase (http://www.shigen.nig.ac.jp/rice/oryzabase/, Yamazaki et al. 2010), the new gene name BHC (BLACK HULL C) is assigned, and Ph is a gene synonym. In this study, the gene symbol Ph is used for its association with phenol reaction and its gene product. Xanthomonas oryzae pv. oryzae (Xoo) is a gram-negative bacterium. It is among the most severe diseases of rice, causing sharp reductions in yield. The dominant allele Xa1 at the XA1 locus is resistant to the Xoo Japanese race I (T7174). The recessive allele xa1 is susceptible to it.
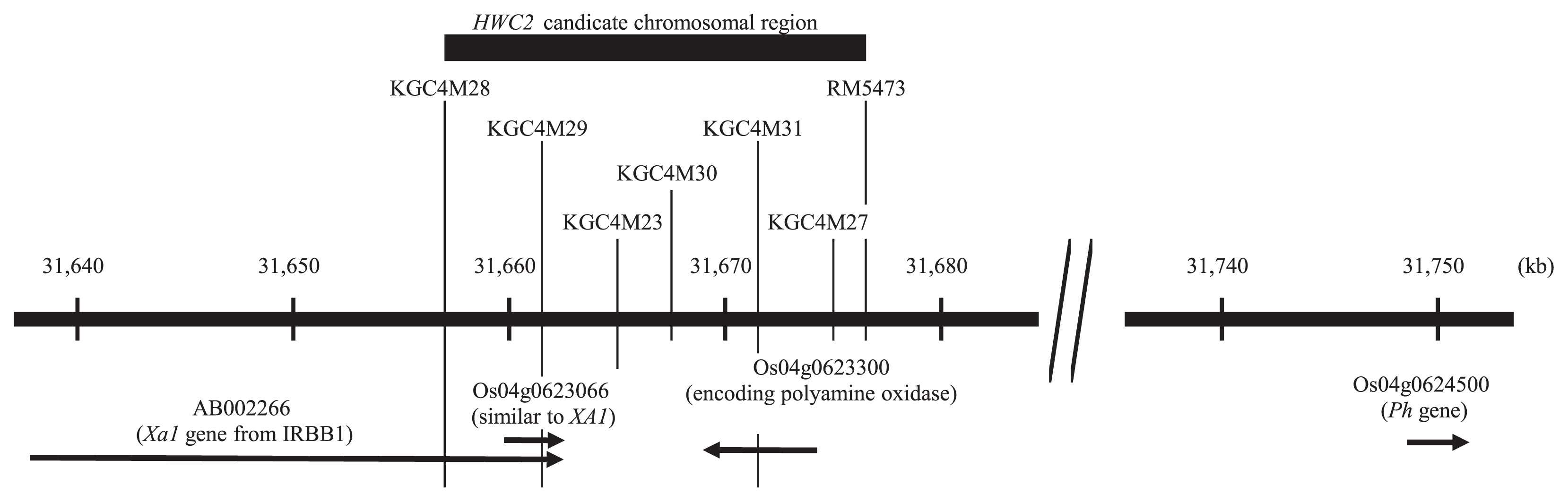
Candidate chromosomal region of HWC2, location of AB002266, Os04g0623066, Os04g0623300 and Os04g0624500 in Os-Nipponbare-Reference-IRGSP-1.0, based on RAP-DB (http://rapdb.dna.affrc.go.jp/). AB002266 is a Xa1 gene from a rice accession IRBB1 (Yoshimura et al. 1998). Os04g0623066 is described as ‘similar to XA1’, and transcribed as AK105096, which corresponds to fourth exon in AB002266. Os04g0624500 is identical to Ph gene (Yu et al. 2008).
In an earlier study (Kuboyama et al. 2009), we conducted high-resolution mapping of HWC2, narrowing down the area of interest to 19 kb. We also performed haplotype analysis around the HWC2 locus and identified the linkage disequilibrium around this locus, which is probably related to regional adaptability and resistance against biotic stress such as bacterial blight, blast, and gall midge. We used only 33 cultivars. Therefore, the breeding significance of haplotypes around HWC2 (HWC2 haplotypes) and varietal differentiation have not been discussed well. For the present study, we extended the HWC2 haplotype analysis to two sets of core collections: the World Rice Core collection (WRC) by Kojima et al. (2005) and the Japanese Rice mini Core collection (JRC) by Ebana et al. (2008). WRC comprises 69 accessions based on a genome-wide RFLP polymorphism survey of 332 accessions of cultivated rice. Actually, WRC includes only two Japanese accessions. The distribution of the Hwc2-1 gene in Japan is also our interest. It could contribute to rice breeding in Japan. Therefore, WRC is insufficient for us. JRC comprises 50 accessions based on a genome-wide SSR polymorphism survey of 236 Japanese landrace accessions. Using the two core collection sets encompassing the wide genetic diversity of rice, we were able to study the distribution of this gene effectively. Because many scientists use these collections (e.g. WRC—Fujino and Sekiguchi 2011, Kanemura et al. 2007, Kusano et al. 2007, Ueno et al. 2009; JRC—Fujino and Sekiguchi 2011, Hori et al. 2009, Nagasaki et al. 2010, Ueno et al. 2009), information related to these collections has accumulated.
Ebana et al. (2010) used most accessions in the two collections to ascertain the DNA sequence diversity and population structure of rice using 4,357 SNPs. The genetic relation among the accessions in the two collections was evaluated using the same criteria. Accessions were classified into Temperate Japonica (TeJ), Tropical Japonica I (TrJ_I), Tropical Japonica II (TrJ_II), Tropical Japonica III (TrJ_III), Indica I (Ind_I), Indica II (Ind_II), Indica III (Ind_III), intermediates such as TrJ_I/Ind_II, which means intermediate between TrJ_I and Ind_II, and mixtures. However, some accessions not used by Ebana et al. (2010) might be important for our study.
In this study, first we applied 40 Insertion/deletion (Indel) markers, most of which were used by Ichitani et al. (2011) for classifying 16 cultivars into temperate Japonica, tropical Japonica, indica, and aus, to ascertain the genetic relation among all accessions in WRC and JRC. Second, we extended the HWC2 haplotype analysis to the two sets of core collections. We also examined the respective reactions to phenol and Xoo Japanese race I to estimate the genotypes of Ph and XA1 loci. Then, we discussed the relation between varietal differentiation and HWC2 haplotypes, and its breeding significance.
WRC and JRC were provided by the National Institute of Agrobiological Sciences (NIAS), Japan. Also, to Table 4 in Kojima et al. (2005), WRC12 was added. Then WRC54 was removed from it (for details, NIAS Genebank database page: https://www.gene.affrc.go.jp/databases-core_collections_wr.php#note02_a). The cultivar name of WRC12 and its varietal group in Ebana et al. (2010) differed from those in the NIAS Genebank database page. All accessions were grown at the experimental farm of the Faculty of Agriculture, Kagoshima University. Each accession showed uniform appearance, suggesting that it was genetically fixed. The Hwc1-1 carrier Jamaica was also grown similarly. The core collection accessions were crossed to Jamaica as a pollen donor. To synchronize the heading dates among core collection accessions and Jamaica, Jamaica was sown and transplanted with an approximately 5-day interval because it is known empirically that Jamaica is weakly photoperiod-sensitive, and that a delay in the sowing date can lead to a delay in the heading date. The core collection accessions were emasculated by soaking their panicles in hot water at 43°C for seven minutes. At least one panicle was left without pollination to confirm that emasculation was complete. After harvest, hybrid seeds were dried at 50°C for five days to break dormancy. They were sown on Petri dishes containing 5-mm-deep tap water and were left in the dark at 28°C for five days. The genotypes of each cultivar were estimated as Hwc2-1/Hwc2-1 if all hybrid plants showed root elongation inhibition (usually less than 1 cm) and hwc2-2/hwc2-2 if they showed normal root elongation (usually more than 5 cm) (Ichitani et al. 2001, Saito et al. 2007) with visual observation. Most hybrid combinations were planted along with parental accessions at the experimental farm of the Kagoshima University to ascertain whether the judgment of HWC2 genotypes immediately after germination was correct, or not.
DNA marker analysisDNA was extracted from one plant for one accession using the process described by Dellaporta et al. (1983), with some modifications. For varietal classification, Ichitani et al. (2011) classified rice cultivars based on 39 Indel markers. Our preliminary analysis using 48 accessions indicated that most markers were di-allelic or tri-allelic, and that they were applicable to all accessions using the same strategy as that reported by Ichitani et al. (2011). However, a marker C52409 had more than four alleles, some of which were difficult to distinguish from one another. Most cultivars classified as Japonica by Ebana et al. (2010) had the same banding patterns as those of Nipponbare (WRC01), a typical temperate Japonica cultivar, whereas most cultivars classified as Indica showed banding patterns similar to those of Kasalath (WRC02), a typical aus cultivar. The migration distances of bands of some Indica cultivars differed slightly from that of Kasalath. These ‘Kasalath-like’ type and Nipponbare type were mutually distinct. For this study, Kasalath type and Kasalath-like type were grouped as the same type. To avoid reduction in information around the C52409, C52190 (http://rgp.dna.affrc.go.jp/E/publicdata/caps/index.html) was added to the present study. E1828 on chromosome 9 was reported to be tri-allelic (Ichitani et al. 2011). Our preliminary analysis revealed that these DNA markers have more than four alleles, which were difficult to distinguish from each other. Therefore, we used a closely linked Indel marker KGC9M8 (Ichitani et al. 2014) instead of E1828. The 40 DNA Indel markers in Supplemental Table 1 were used for this study. For HWC2 haplotypes, 15 DNA markers (Supplemental Table 2) were used. The PCR conditions for Indel and CAPS markers used for this study were the following: 95°C for 5 min, 40 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, with subsequent final extension of 72°C for 1 min, except that the annealing temperature of KGC4M29 was 60°C instead of 55°C. The PCR conditions used for a dCAPS marker KGC4M31 were the following: 95°C for 10 min, 10 cycles of 94°C for 30 s, 55°C for 2 min, and 72°C for 30 s, followed by 25 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, with final extension of 72°C for 1 min. The annealing temperature of another dCAPS marker KGC4M30 was 50°C instead of 55°C. The PCR mixture (5 μl) contained ca. 10 ng template DNA, 200 μM of each dNTP, 0.2 μM of primers, 0.25 units of Taq polymerase (AmpliTaq Gold; Applied BioSystems, CA, USA), and 1 × buffer containing MgCl2. The PCR products were resolved using electrophoresis in 10% (29:1) polyacrylamide gel, followed by ethidium bromide staining and viewing under ultraviolet light irradiation. We obtained good resolution of polymorphic Indel bands by running the electrophoresis until xylen cyanol migrated 5–5.5 cm from the origin. When analyzing CAPS and dCAPS, we obtained good resolution of polymorphic bands by continuing electrophoresis until xylen cyanol migrated 3–4 cm from the origin.
Phenol reactionTo confirm the phenol reaction in each accession, one or two seeds were soaked in 3% phenol solution for three days and were dried overnight. Accessions with hulls that turned black carried the Ph allele. Those with hulls which remained the original color carried the ph allele.
Bacterial race, its inoculation and scoringTo ascertain the relation between HWC2 haplotype and resistance to Xoo, we infected the two core collections with Xoo Japanese race I (T7174). Carriers of resistant allele Xa1 at XA1 locus are highly resistant to T7174, although the carriers of the resistant allele Xa7 at XA7 locus located on chromosome 6 are also highly resistant to the race (Ogawa et al. 1991). As discussed below, however, if cultivars sharing a certain haplotype surrounding HWC2 locus showed resistance to this race, it could be a sign of carrying Xa1 allele. The Xoo inoculum was prepared and cultured using potato semi-synthetic agar media (Wakimoto 1954) and incubated at 28°C for 48 hr. The inoculum was then diluted with distilled water. The absorbance was adjusted to A = 0.05 (620 nm) using a spectrophotometer. This value corresponds to concentration of about 108 colony forming units per milliliter (cfu/ml), which normally provides optimum conditions. Inoculation was performed using the clipping method according to procedures described by Kauffman et al. (1973) in late August in 2008, when many accessions were in the booting stage or later stage. Scoring of resistance was done by measuring the lesion length of three infected leaves with a ruler 18 days after inoculation of the Xoo pathogen.
Statistical analysisTwo classification methods were applied. One was based on the frequency of Japonica specific alleles (Fj), as proposed by Lu et al. (2009) and pursued in later work by Xiong et al. (2010). As explained in the Results section, accessions were classified roughly into two groups. Accessions in one group shared the same banding patterns as Nipponbare at most loci. The same banding pattern as that of Nipponbare was coded allele A. Accessions in the other group shared banding patterns that differed from those of Nipponbare at most loci. Most of these were shared by Kasalath. The same banding pattern as that of Kasalath was coded as allele B. The other banding patterns shared by neither Nipponbare nor Kasalath were coded as C, D, etc., according to frequencies at the respective loci. At KGR4M30 and S0494 loci, Nipponbare and Kasalath shared the same and most-frequent banding patterns. The second most frequent bands were coded B at these loci. In this study, Fj was calculated as the frequency of A alleles.
The other classification was shown to have genetic similarity using the Jaccard coefficient (Jaccard 1908). Each fragment size was treated as a unique characteristic and was scored as present (1) or absent (0). The data matrix was used to calculate genetic similarities using the Jaccard (1908) coefficient. Statistical methods of two types classifying accessions were applied: (1) unweighted pair-group method with an arithmetic mean (UPGMA) cluster analysis and (2) principal component analysis (PCA). Both analyses were conducted using software (PAST Version 3.11; Hammer et al. 2001).
SNP data collection around HWC2CAPS and dCAPS markers have been developed by finding the SNPs distinguishing an Hwc2-1 carrier Nipponbare from the three hwc2-2 carriers (Kasalath, Guangluai 4 and Jamaica) (Kuboyama et al. 2009). For more SNP information for both cultivated and wild rice accessions around HWC2 locus, we downloaded a SNP file of 1,083 cultivated rice accessions and 446 wild rice (O. rufipogon) accessions for chromosome 4 ‘Cul_Wild_1529lines.chr4’ from the Rice Haplotype Map Project database (http://www.ncgr.ac.cn/RiceHap3), which has contributed to revealing the origin of cultivated rice (Huang et al. 2012). Because this file is too large (2.14 Gb) and difficult to handle, we used software (Div 1.10a; http://www.vector.co.jp/soft/win95/util/se088352.html) to divide it into subfiles (ca. 30 Mb each) comprising 10,000 SNP sites × 1,529 accessions. Subfiles were read using a spreadsheet program (Office Excel 2007; Microsoft Corp., Redmond, WA, USA).
Results of Fj show that accessions were generally classifiable into two groups (Supplemental Fig. 1). One group was composed of Indica in Ebana et al. (2010) and in this study (see below). The other group was composed of Japonica. Mixture, TrJ_I/Ind_II, and TrJ/Ind_III in Ebana et al. (2010) were scattered between Indica and Japonica. WRC12, categorized as TrJ_III, showed Fj of 0.125. WRC43, categorized as Ind_II/TeJ, showed Fj of 1.000. These Fj values were consistent with the results found for UPGMA and PCA (see below).
The relations among both core collections were also revealed by UPGMA cluster analysis (Fig. 2). All accessions were classified clearly into two major groups respectively corresponding to Japonica and Indica in Ebana et al. (2010). Each group was divided by subgroups based on the clusters.

Relation of 119 rice accessions using 40 Indel markers, depicted based on results of UPGMA cluster analysis. Origin and classification by Ebana et al. (2010), and Fj are also shown.

Relation of 119 rice accessions using 40 Indel markers, depicted based on results of UPGMA cluster analysis. Origin and classification by Ebana et al. (2010), and Fj are also shown.
The Japonica group was divided into four subgroups: Japonica_a (J_a) consisted of 33 accessions including Nipponbare. Thirty accessions were from Japan. The other three were from China, Laos, and Vietnam. Fj was 0.90–1.0. They shared A allele at 29 loci. Among the 28 accessions examined by Ebana et al. (2010), 24 were categorized as TeJ. Nipponbare (WRC01) with Fj equal to 1 are generally regarded as temperate Japonica. Therefore, it is natural that accessions with high Fj were categorized as temperate Japonica. The other four accessions were categorized as one TrJ_I, one TrJ_I/TeJ, one TrJ_II/TeJ, and one Ind_II/TeJ. Therefore, accessions in subgroup J_a were categorized as temperate Japonica. Japonica_b (J_b) consisted of 15 accessions. They were all from Japan. The JRC lines in this clade were upland rice (Ebana et al. 2008). Accessions in J_b were categorized as TrJ_I or TrJ_I/TeJ. Therefore, accessions in subgroup J_b were classified as tropical Japonica. Their Fj were 0.775–0.90. They share A allele at 29 loci. At some loci, B alleles were prevalent: S5756 (12/15), S20234 (10/15), C51124 (15/15), KGS0342 (15/15), and KGS1797 (15/15). Japonica_c (J_c) consisted of nine accessions. In Ebana et al. (2010), they were categorized as two TeJ, three TrJ_I, two Tr_II, one TrJ_III, and one mixture. They originated from various countries: five were from Southeast Asia; two were from Japan; one was from the United States of America; and one was from Brazil. Their Fj were 0.750–0.825 (Fig. 2). They shared the A allele at 19 loci. At some loci, B alleles were prevalent: S5756 (5/9), C61009 (9/9), KGR4M30 (7/9), C30372 (5/9), S20234 (6/9), KGS0342 (7/9), KGS1797 (6/9), and RZ869 (5/9). At KGC9M8 locus, C allele was the most popular allele in this subgroup. Most accessions belonged to tropical Japonica. Therefore, accessions in J_c were categorized as tropical Japonica. Furthermore, WRC24, WRC44, WRC53, WRC55, and JRC41 were distant from the other accessions. Their Fj were 0.50–0.70, which was smaller than the other Japonica groups. In a report by Ebana et al. (2010), WRC53 and WRC55 were categorized as mixtures, with WRC24 as Trj_I/Ind_III and WRC44 as TrJ/In_III. For this study, these cultivars were categorized as Intermediate (Int).
Indica group was divided into three subgroups. Indica_a (I_a) consisted of 34 accessions. Fj were 0.100–0.250. Among the 32 accessions examined by Ebana et al. (2010), 11 and 20 accessions were categorized respectively as Ind_II and Ind_III, corresponding to indica in a description by Garris et al. (2005). Therefore, I_a could be categorized as indica. Resolution by 40 Indel loci was insufficient to classify this group into Ind_II and Ind_III. WRC12 was categorized as Tr_III in Ebana et al. (2010). However, it was categorized as Indica (I_a) for this study. Accessions in I_a shared B alleles at 12 loci. A allele-prevalent loci of I_a were: C52190 (32/34), S20049 (20/34), KGS0049 (22/34), E30287 (22/34), E12196 (24/34), and E4443 (34/34). Indica_b (I_b) consisted of 19 accessions including Kasalath. Fj were 0.05–0.250. They all were categorized as Ind_I in Ebana et al. (2010), which corresponded to aus in Garris et al. (2005) (Ebana et al. 2010). These accessions shared B alleles at 22 loci. At some loci, A alleles were prevalent: KGR4M30 (11/19), KGS0049 (12/19), KGR4M43 (10/19), KGS0494 (19/19). Also, WRC21, WRC22, WRC23, and WRC42 were distant from the other accessions. Their Fj were 0.275–0.500, which was larger than those of other Indica groups. Actually, WRC21 and WRC42 were categorized respectively as Ind_I and Ind_III in Ebana et al. (2010). In addition, WRC22 and WRC23 were categorized as TrJ_I/Ind_III. In this study, these cultivars were categorized as Int. As a whole, classification by the 40 Indel markers were consistent with that by Ebana et al. (2010) using 4,357 SNPs. Accessions not analyzed in Ebana et al. (2010) were able to be categorized as follows: JRC28, JRC40, JRC53, JRC39, and JRC50 were categorized as J_a (temperate Japonica). JRC41 was categorized as Int. JRC42 and JRC 21 were categorized as I_a (indica) (Fig. 2).
Based on PCA, accessions were classified into three major groups by the first two principal components (Supplemental Fig. 2). The variances explained by the first and second principal components were, respectively, 63.3% and 5.64%. Accessions in J_a, J_b, and J_c formed one group. Accessions in I_a and those in I_b formed different groups. Accessions in the same groups in Fig. 2 were surrounded by dotted circles. The nine accessions outside of the circles were categorized as Int in Fig. 2.
Distribution of HWC2 and Ph genesAll 119 accessions were genotyped for HWC2 and Ph loci (Table 1): 37 accessions carried Hwc2-1 allele, whereas 82 accessions carried hwc2-2 allele. In total, 30 out of 33 J_a, equivalent to temperate Japonica, carried Hwc2-1. The remaining seven accessions carrying Hwc2-1 were two J_b, two J_c, two I_a, and one I_b. The opposite situation was found for hwc2-2 carriers. 20 out of 24 J_b and J_c, which are equivalent to tropical Japonica, and 50 out of 53 I_a and I_b, which are equivalent to Indica, accessions carried hwc2-2. Regarding Ph locus, 57 accessions carried Ph allele whereas 62 accessions carried ph allele. The phenol reaction result suggests that the genotypes of Ph locus corresponded to DNA marker-based varietal classification. The distribution of the Ph allele was skewed toward Indica accessions whereas that of ph allele was skewed toward Japonica accessions: 41 out of 53 Indica accessions carried Ph allele whereas 46 out of 57 Japonica accessions carried ph allele. Strong association between HWC2 and Ph was also observed (Table 1, χ2 for independence = 43.951, P < 0.001). 36 out of 37 Hwc2-1 carriers had ph allele, whereas 56 out of 82 hwc2-2 carriers had Ph allele.
| Varietal group | Allele at HWC2 locus | Allele at Ph locus | Sum | |
|---|---|---|---|---|
| Ph | ph | |||
| Japonica_a (J_a) | Hwc2-1 | 0 | 30 | 30 |
| Japonica_b (J_b) | 0 | 2 | 2 | |
| Japonica_c (J_c) | 0 | 2 | 2 | |
| Indica_a (I_a) | 0 | 2 | 2 | |
| Indica_b (I_b) | 1 | 0 | 1 | |
| Intermediate (Int) | 0 | 0 | 0 | |
| Sum | 1 | 36 | 37 | |
| Japonica_a (J_a) | hwc2-2 | 1 | 2 | 3 |
| Japonica_b (J_b) | 9 | 4 | 13 | |
| Japonica_c (J_c) | 1 | 6 | 7 | |
| Indica_a (I_a) | 27 | 5 | 32 | |
| Indica_b (I_b) | 13 | 5 | 18 | |
| Intermediate (Int) | 5 | 4 | 9 | |
| Sum | 56 | 26 | 82 | |
The banding patterns of 14 DNA markers around the HWC2 locus (Supplemental Table 2) of 119 accessions genotypes at Ph locus are depicted in Fig. 3. Regarding IBA44, the carrier of the second frequent allele was confined to two accessions (WRC2 and WRC37). Therefore, it was excluded from further study. Regarding the other loci, carriers of the second frequent allele were more than 13. Some accessions produced two banding patterns at KGC4M29: one was digested with RsaI, the other was not. In our earlier paper (Kuboyama et al. 2009), we reported that the Vietnamese cultivar Te-Tep produced such a banding pattern. Such accessions shared the similar HWC2 haplotypes (see below). Although the correct chromosomal location of the ‘duplicated’ KGC4M29 locus remains unknown, the two bands were treated as the third banding pattern based on the assumption that the original and the duplicated KGC4M29 loci were linked closely to one another. When using 14 markers, the 119 cultivars were classified into 44 haplotypes. The Ph locus is located between S13714 and KGC4M52. When the genotype of Ph locus was added, they were classified into 50 haplotypes.
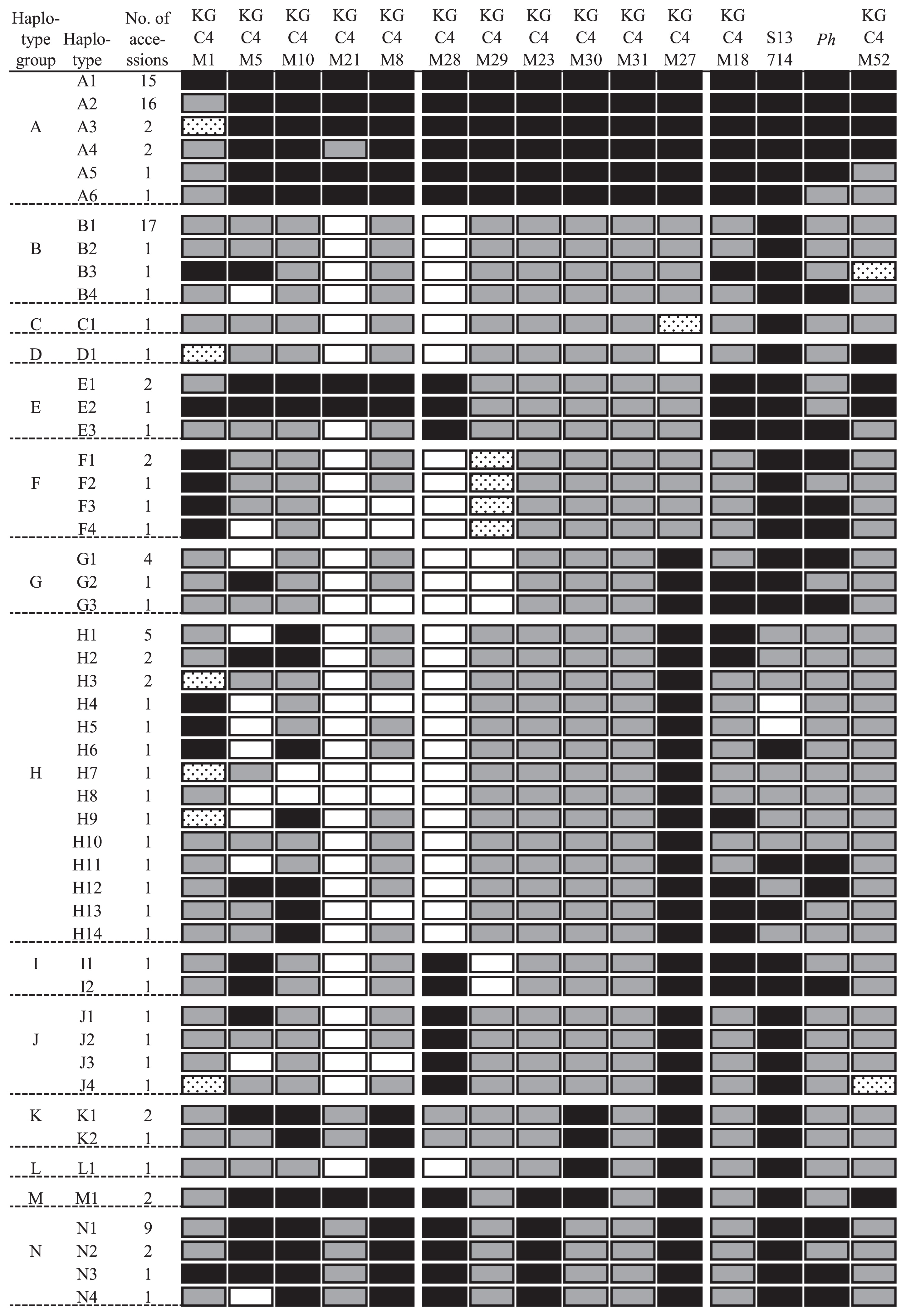
Haplotype diversity around Hwc2 locus evaluated using the 14 DNA markers closely linked with HWC2 locus and the genotype of Ph locus. The same banding pattern as that of Nipponbare is denoted by solid rectangles. The most frequent patterns other than those of Nipponbare are denoted by shaded rectangles. The other patterns are denoted by dotted rectangles. Open rectangles signify that no band appeared. Regarding the Ph locus, ph and Ph alleles are denoted respectively as solid and shaded rectangles. The haplotype groups characterized by the six DNA markers closely linked with HWC2 locus, KGC4M28, KGC4M29, KGC4M23, KGC4M30, KGC4M31, and KGC4M27, are denoted by alphabet letters. The haplotypes characterized by the 14 DNA markers and Ph locus are denoted by alphabet letters with Arabic numerals.
The Hwc2-1 carriers belonged to six haplotypes. They showed polymorphism at KGC4M1, KGC4M21, Ph, and KGC4M52 loci. Regarding the other loci, they shared the same banding patterns as Nipponbare (WRC01). The A1 and A2 haplotypes respectively comprised 15 and 16 accessions: they were much more frequent than the other Hwc2-1 haplotypes. The hwc2-2 carriers were classified into 44 haplotypes. Among them, two haplotypes with many accessions were present. Haplotype B1 comprised 17 accessions, many of which originated from Japan, and which belonged to J_b (tropical Japonica) or I_a (indica). Haplotype N1 comprised 9 accessions, most of which originated from Southeast Asia, and which belonged to J_a, J_c (tropical Japonica) or I_b (aus). The other hwc2-2 haplotypes were composed of one to five accessions.
To examine the HWC2 candidate chromosomal region specifically, haplotype analyses were conducted of six markers within the candidate region of HWC2 (Kuboyama et al. 2009): KGC4M28, KGC4M29, KGC4M23, KGC4M30, KGC4M31, and KGC4M27. The relation among HWC2 haplotypes of the six DNA markers is presented in Fig. 4. Hereinafter, the haplotype of the six DNA markers, into which the haplotypes of the 14 DNA markers and Ph locus were grouped, is named the ‘haplotype group’. Hwc2-1 carriers formed one haplotype group, separated by the haplotype groups of hwc2-2 carriers. Nipponbare type alleles at KGC4M29 and KGC4M31 loci completely corresponded to Hwc2-1 allele, which is consistent with the previous study (Kuboyama et al. 2009). The hwc2-2 carriers were classified into 13 haplotype groups: haplotype groups B, H, and N were composed of more than 12 accessions. The other haplotype groups were composed of fewer than seven accessions. Haplotype group B was composed of ten I_a accessions, eight J_b, one J_c, and one Int accessions. They shared the same banding patterns from KGC4M10 to KGC4M31. Haplotype group B was the same as that of the Xa1 carrier ‘Java14’ (Kuboyama et al. 2009, Ogawa et al. 1991): 16 accessions showed lesion length shorter than 1.0 cm, indicating high resistance, suggestive of a Xa1 carrier. When evaluated using 14 DNA markers and Ph genotypes, accessions in the haplotype group H were divided into 14 types (Fig. 3). Haplotype group H was composed mainly of I_a and I_b accessions. Haplotype group N was composed of five I_b, one Int, and seven Japonica cultivars that originated from Southeast Asia (two J_a and five J_c). They shared the same banding patterns from KGC4M10 to S13714. Accessions in Haplotype F were characterized by the unique banding pattern at the KGC4M29 locus: two bands appeared as presented above. They shared the same haplotype with the Xa1 carrier Te-Tep (Kuboyama et al. 2009, Ogawa et al. 1991). All five accessions showed lesion length shorter than 1.0 cm, indicating high resistance, suggestive of carrying Xa1 allele. Most accessions in the other haplotype groups showed lesion length longer than 1.0 cm, indicating that they carried susceptible allele xa1 at the Xa1 locus.
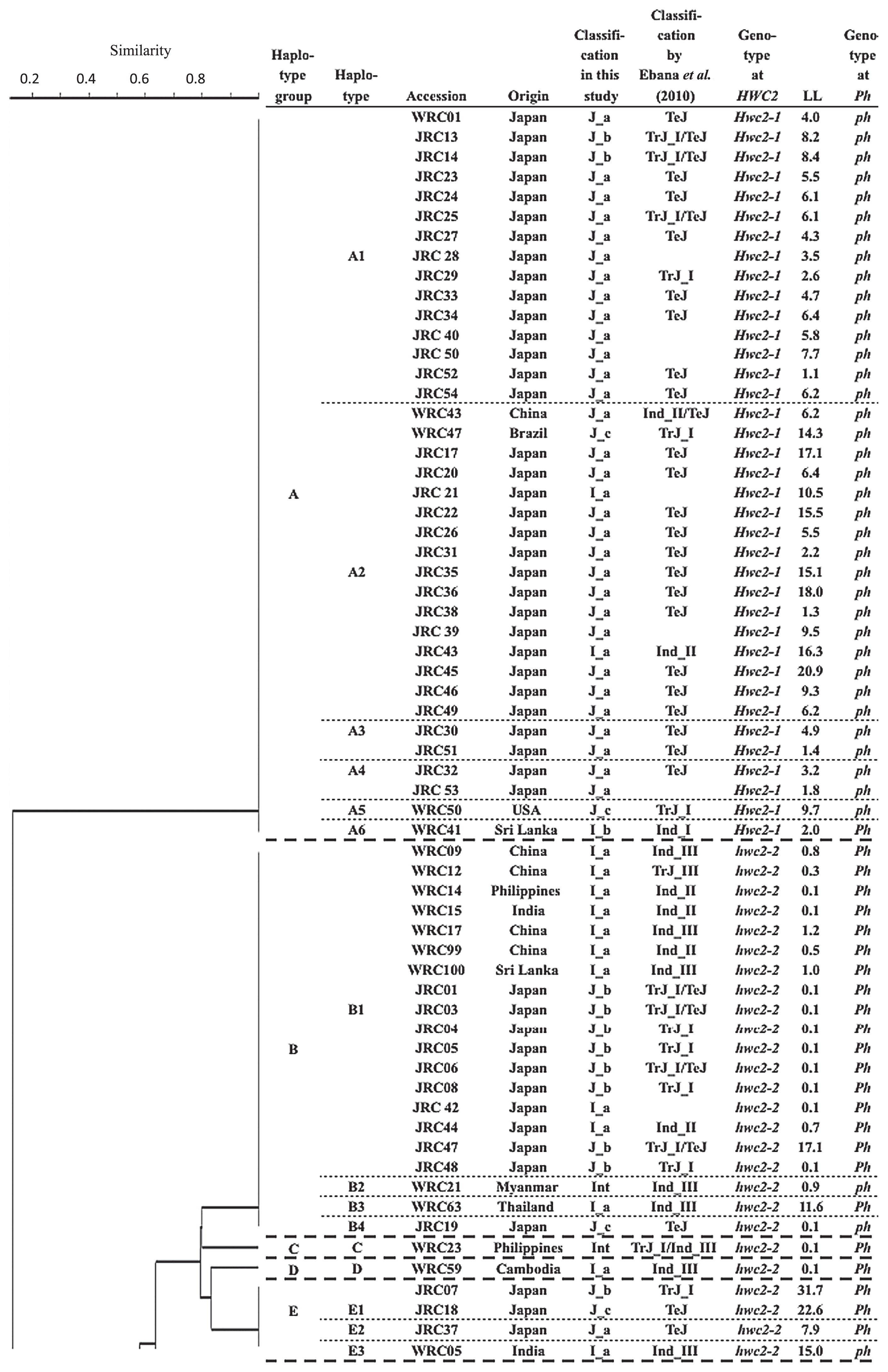
Relation of 119 rice accessions using six DNA markers linked closely with HWC2 locus, as depicted by UPGMA cluster analysis. The ‘haplotype group’ characterized by the six DNA markers, ‘haplotype’ characterized by the 14 DNA markers and the genotype of Ph locus (Fig. 3), origin, classification in this study, that by Ebana et al. (2010), genotypes at the HWC2 locus, average lesion length (cm) of three leaves inoculated with Xoo Japanese race I (LL), the genotypes at the Ph locus are also shown.
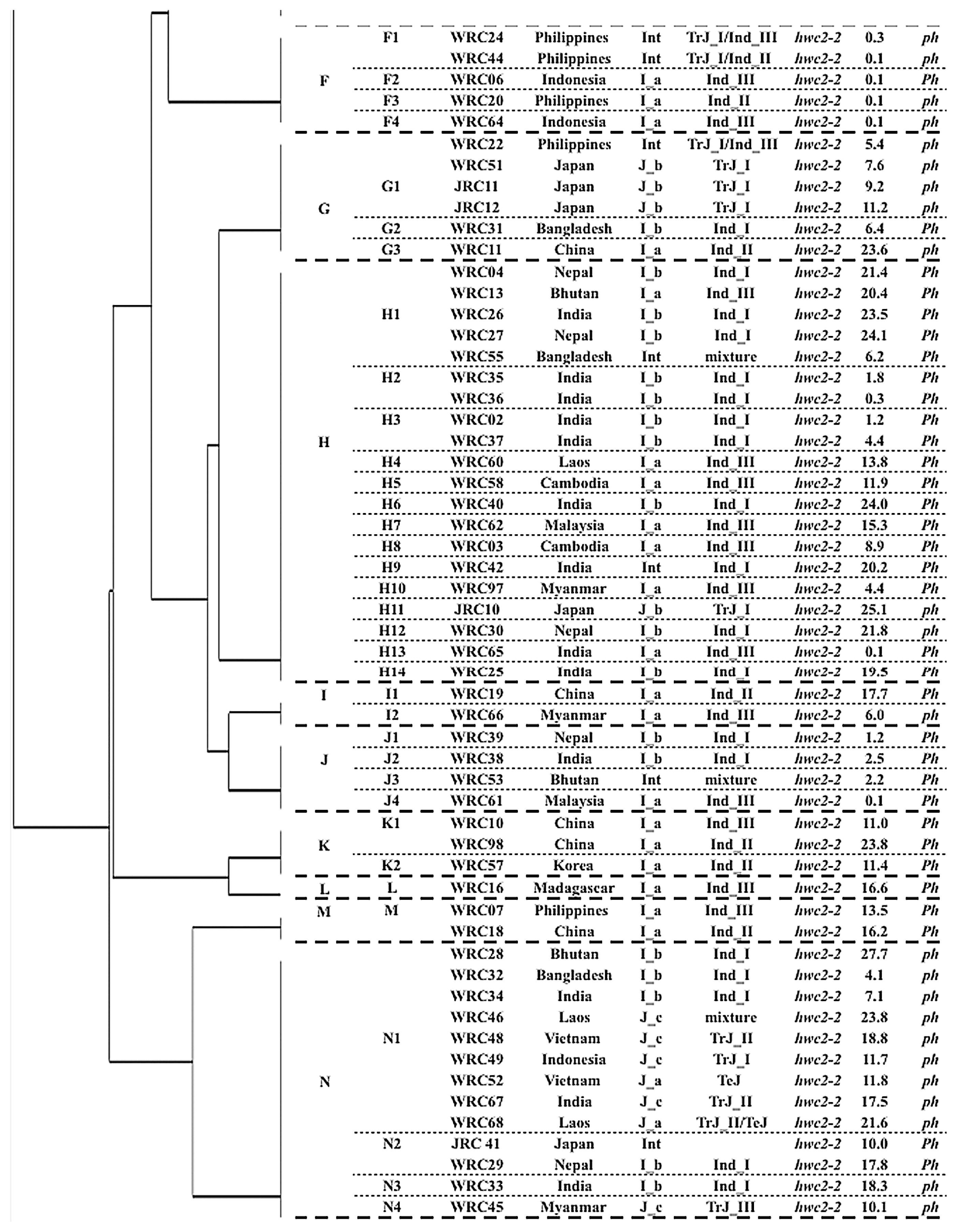
Relation of 119 rice accessions using six DNA markers linked closely with HWC2 locus, as depicted by UPGMA cluster analysis. The ‘haplotype group’ characterized by the six DNA markers, ‘haplotype’ characterized by the 14 DNA markers and the genotype of Ph locus (Fig. 3), origin, classification in this study, that by Ebana et al. (2010), genotypes at the HWC2 locus, average lesion length (cm) of three leaves inoculated with Xoo Japanese race I (LL), the genotypes at the Ph locus are also shown.
Among SNPs detected by CAPS and dCAPS markers, the one recognized by KGC4M31 was detected in ‘Cul_Wild_1529lines.chr4’. Results of the two core collections show that adenine (A) at 31,671,1271 on the Os-Nipponbare-Reference-IRGSP-1.0 was shared by Hwc2-1 carriers, and that thymine (T) at the same position was shared by hwc2-2 carriers. Among 308 O. rufipogon accessions in which DNA sequence of KGC4M31 SNP site was read, only one accession (W3045) carried A-type. The other accessions carried T-type (Table 2). Among 480 cultivated rice accessions, 134 carried A-type; 346 carried T-type.
| Base | Accessions in this studya | Accessions in Huang et al. (2012) | ||
|---|---|---|---|---|
| Hwc2-1 carrier | hwc2-2 carrier | O. rufipogon | O. sativa | |
| A | 37 | 0 | 1 | 134 |
| T | 0 | 82 | 307 | 346 |
| Undetermined | 0 | 0 | 138 | 603 |
In this study, the distribution of the alleles of HWC2, one causal gene of a hybrid weakness phenomenon in rice, and the haplotype surrounding this gene was surveyed using two sets of core collections comprising 119 accessions from the standpoint of varietal differentiation and rice breeding history. Not all accessions had been analyzed using the same DNA markers. Seven accessions had not been categorized into varietal groups. Therefore, all accessions in the two core collections were analyzed using 40 published Indel markers. The classification used for this study is almost consistent with that described by Ebana et al. (2010) using 4,357 SNPs. Indel polymorphism is expected to be rarer than with SNP. For example, Xu et al. (2012) sequenced 40 cultivated rice accessions including tropical Japonica, temperate Japonica, aromatic, indica, aus, groups III and IV proposed by Glaszmann (1987), and ten wild rice accessions including five O. rufipogon and five O. nivara, to 15 × coverage using Illumina GA2 instruments. They identified 6,496,456 high-quality SNPs, 808,000 small Indels ranging from 1 bp to 5 bp. For larger Indels, they pooled accessions from the same groups, which yielded > 80 × raw coverage of the reference rice genome. No detailed data were included in their report. Judging from the supplemental data, which can be downloaded from ftp://rice:ricedownload@public.genomics.org.cn/BGI/rice, Indels of 10–100 bp were estimated as ca. 340,000. Those larger than 100 bp were ca. 94,700. Therefore, large Indels are thought not to be appropriate for intragroup diversity. Nevertheless, these results suggest that large Indels might result from infrequent but probably irreversible and robust mutation, and that they might be shared and retained by each of major groups: temperate Japonica, tropical Japonica, aromatic, indica, and aus. Therefore, a few large Indels detecting the allelic difference in this study might reflect varietal differentiation such as Indica–Japonica, temperate Japonica–tropical Japonica, and aus–indica with high accuracy.
Results of the distribution of HWC2 and its haplotype analysis are summarized as follows. (1) Hwc2-1 carriers were prevalent among temperate Japonica, but rare among the other varietal groups. Strong association between HWC2 and Ph was also observed. (2) Hwc2-1 carriers shared one HWC2 haplotype group; hwc2-2 carriers had 13 HWC2 haplotype groups. The three large hwc2-2 haplotype groups were characterized by different varietal groups. Each of the three hwc2-2 haplotype groups comprised plural varietal groups. (3) Resistance to Xoo Japanese race I was related to some haplotype groups to which carriers of the Xa1 allele, conferring resistance to this race, belonged.
Accessions in the same varietal group shared the same banding pattern at most Indel loci scattered in the rice genome. Therefore, before starting this study, we expected that accessions in one group tended to share one HWC2 haplotype, and that this haplotype could be a varietal group indicator. This idea is true for almost all J_a (Fig. 4): 30 out of 33 accessions carried Hwc2-1 allele and the same haplotype group. However, the other varietal groups had more than three HWC2 haplotype groups: I_a had as many as 12 HWC2 haplotype groups. From the viewpoint of accessions comprising each HWC2 haplotype group, the popular HWC2 haplotype groups comprised different varietal groups. These results indicate that the relation based on the HWC2 haplotypes group differs completely from that based on the DNA polymorphism of the whole genome. Diversity of Hwc2-1 and hwc2-2 haplotypes is discussed separately.
The skewed distribution of Hwc2-1 gene in temperate Japonica is consistent with results reported for previous studies (Kuboyama et al. 2009, Sato and Hayashi 1983, Sato and Morishima 1987, 1988). The reason for the prevalence of Hwc2-1 in temperate Japonica cultivars remains unknown. One plausible idea is that genes under selection are linked closely to Hwc2-1 gene. The candidate of such a gene is Ph. HWC2 and Ph are linked closely (Ichitani et al. 2001). Ph gene encodes polyphenol oxidase (PPO) (Yu et al. 2008). It is identical to Os04g0624500. The physical distances between Ph and most closely linked DNA markers to HWC2 (Kuboyama et al. 2009) are 75–93 kb (Fig. 1). Yu et al. (2008) demonstrated that the positive Ph allele is responsible for darker grain color after harvest. They proposed that Japonica cultivar grains are refractory to discoloration during storage, which might be an aesthetic appeal, because of which inactive ph allele is more likely to be artificially selected. The possible reason for Indica cultivars maintaining Ph allele is that plant PPOs are reportedly associated with disease resistance, and that agriculture in the tropical and subtropical zone still assigns a premium to disease resistance, much like the case in the wild. The prevalence of Hwc2-1 and ph allele in temperate-Japonica is explainable by the two hypotheses. (1) The mutation causing Hwc2-1 allele might occur in the carriers of ph allele in the prototype of temperate Japonica. (2) The mutation from hwc2-2 to Hwc2-1 and that from Ph to ph occurred before varietal differentiation. Another possibility is that Hwc2-1 in itself, aside from causing hybrid weakness in the presence of Hwc1-1 gene, has a pleiotropic effect on the adaptability to rice growing conditions in eastern Asia, where temperate Japonica is frequently used. The identification of HWC2 gene is underway.
Carriers of hwc2-2 were classified into 13 haplotype groups (Figs. 3, 4). As discussed above, the three large hwc2-2 haplotype groups were characterized by different varietal groups. Each of the three hwc2-2 haplotype groups comprised plural varietal groups. The reason is explainable in light of two hypotheses: (1) Introgression of hwc2-2 haplotype group occurred from one varietal group to another. (2) Genetic diversity before varietal differentiation has been shared in different varietal groups. Both hypotheses suggest artificial or natural selection of gene(s) along with some hwc2-2 haplotypes. One candidate of such a gene is the Xoo resistance gene. HWC2 haplotype groups have a strong association with the reaction to Xoo Japanese race I, a good indicator of genotype of XA1, a candidate gene of HWC2. Java14, a carrier of the resistant allele Xa1 (Ogawa et al. 1991), had the same haplotype as haplotype group B (Kuboyama et al. 2009). Results show that 18 out of 20 accessions in haplotype group B were highly resistant to Xoo Japanese race I, suggesting that they carry Xa1. Actually, Xa1 allele behaves as the dominant gene. Its expression is induced by bacterial inoculation (Yoshimura et al. 1998). Therefore, susceptible accessions can result from a loss of function mutation of Xa1. Other genes for resistance against pests such as blast and gall midge are also located near the HWC2 locus (Kuboyama et al. 2009). Haplotypes H and N, other popular hwc2-2 haplotypes, might carry such resistance gene(s), although they seemed not to carry Xa1. Haplotype group F is characterized by two banding patterns at KGC4M29. Te-tep, another carrier of the resistant allele of Xa1, had the same haplotype (Kuboyama et al. 2009). All five accessions of haplotype group F are resistant to Xoo Japanese race I, suggesting that they carry Xa1. Xa7 at the XA7 locus confers high resistance against Xoo Japanese race I (Ogawa et al. 1991). This gene is located on chromosome 6 (Kaji and Ogawa 1995). According to Sidhu et al. (1978), Lee and Khush (2000), and Khan et al. (2014), the carriers of this gene are limited to a few aus cultivars. The fact that no aus (I_b) accession belongs to Haplotype groups B and F supports the inference that Xoo resistance in the accessions in the Haplotype groups was conditioned by Xa1, not by Xa7.
The genotypes of KGC4M29 and KGC4M31 coincided completely with those for HWC2, consistent with our earlier study (Kuboyama et al. 2009). The restriction enzyme recognition site of KGC4M29 was located in Os04g0623066 encoding protein similar to XA1: Xa1 allele of IRBBI encodes a cytoplasmic receptor-like protein with nucleotide binding sites (NBS) and leucine-rich repeat domains (LRR) (Yoshimura et al. 1998). Its fourth exon, which contains NBS and LRR, corresponds to Os04g0623066 (Fig. 1. RAP-DB: http://rapdb.dna.affrc.go.jp/index.html). The restriction enzyme recognition site of KGC4M31 was in Os04g0623300, encoding polyamine oxidase (RAP-DB) (Fig. 1). Results of that study suggest that Os04g0623066 and Os04g0623300 are candidate genes of HWC2. According to the SNP database in Huang et al. (2012), among 308 wild rice accessions, only one accession (W3045) carried the same SNP as Hwc2-1 carriers at KGC4M31 (Table 2). If the association between HWC2 genotypes and the SNP can be conserved in O. rufipogon, then these data indicate that carriers of Hwc2-1 allele are very rare in O. rufipogon, which is consistent with results reported by Sato and Morishima (1987). Hwc2-1 allele is encompassed by temperate Japonica specific haplotype. Therefore, W3045 might hold some clue to the origin of temperate Japonica. However, some possibility exists of introgression from the cultivated rice into wild rice because 18 bp deletions in the ph allele of most Japonica cultivars were detected in three O. rufipogon accessions (Yu et al. 2008). The chromosomal region containing HWC2 and Ph includes accumulated genetic information related to the birth of temperate Japonica. Furthermore, HWC2 haplotype analysis of O. rufipogon is expected to answer the question of whether the genetic diversity found in hwc2-2 haplotypes had originated from O. rufipogon and selected irrespective of varietal groups, or if it occurred and has since been accumulated during the rice domestication process.
We are grateful to the Genebank of National Institute of Agrobiological Sciences for their kind provision of core collections. This work was supported by JSPS KAKENHI Grant number 24580009 and by the Ministry of Agriculture, Forestry and Fisheries of Japan (Green Technology Project GD-2005), Japan.