Abstract
In the model species Streptomyces coelicolor A3(2), the uptake of chitin-degradation byproducts, mainly N,N′- diacetylchitobiose ([GlcNAc]2) and N-acetylglucosamine (GlcNAc), is performed by the ATP-binding cassette (ABC) transporter DasABC-MsiK and the sugar-phosphotransferase system (PTS), respectively. Studies on the S. coelicolor chromosome have suggested the occurrence of additional uptake systems of GlcNAc-related compounds, including the SCO6005–7 cluster, which is orthologous to the ABC transporter NgcEFG of S. olivaceoviridis. However, despite conserved synteny between the clusters in S. coelicolor and S. olivaceoviridis, homology between them is low, with only 35% of residues being identical between NgcE proteins, suggesting different binding specificities. Isothermal titration calorimetry experiments revealed that recombinant NgcESco interacts with GlcNAc and (GlcNAc)2, with Kd values (1.15 and 1.53 μM, respectively) that were higher than those of NgcE of S. olivaceoviridis (8.3 and 29 nM, respectively). The disruption of ngcESco delayed (GlcNAc)2 consumption, but did not affect GlcNAc consumption ability. The ngcESco-dasA double mutation severely decreased the ability to consume (GlcNAc)2 and abolished the induction of chitinase production in the presence of (GlcNAc)2, but did not affect the GlcNAc consumption rate. The results of these biochemical and reverse genetic analyses indicate that NgcESco acts as a (GlcNAc)2- binding protein of the ABC transporter NgcEFGSco-MsiK. Transcriptional and biochemical analyses of gene regulation demonstrated that the ngcESco gene was slightly induced by GlcNAc, (GlcNAc)2, and chitin, but repressed by DasR. Therefore, a model was proposed for the induction of the chitinolytic system and import of (GlcNAc)2, in which (GlcNAc)2 generated from chitin by chitinase produced leakily, is mainly transported via NgcEFG-MsiK and induces the expression of chitinase genes and dasABCD.
Streptomycetes are multicellular mycelial bacteria that thrive in soil environments as well as in marine and fresh water ecosystems. As producers of a large range of secondary metabolites, including two-thirds of all known antibiotics as well as many anticancer, antifungal and immunosuppressive agents, streptomycetes are of utmost importance for human health, agriculture, and biotechnology (1, 2). Streptomycetes have a saprophytic lifestyle and degrade all naturally occurring biopolymers; therefore, they are a rich source of industrially relevant enzymes (12, 47). These bacteria are major decomposers of chitin, a polymer of beta-1,4-linked N-acetylglucosamine (GlcNAc) units. Complete chitin degradation into GlcNAc and N,N′-diacetylchitobiose ([GlcNAc]2) by streptomycetes requires the production of extracellular chitinases of families 18 and 19 of the glycoside hydrolase (GH) classification (for a review, see [28]), intra- and extracellular N-acetyl-β-D-glucosaminidases of GH families 3 and 20 (15, 33, 44), and the lytic polysaccharide monooxygenase of AA10 (21), the amino acid sequence of which is similar to chitin-binding proteins (29, 38).
The uptake of chitin degradation byproducts was initially studied in Streptomyces olivaceoviridis, which uses PtsC2, the transmembrane enzyme IIC of the phosphoenolpyruvate phosphotransferase system (PTS), and the ATP-binding cassette (ABC) transporter NgcEFG for GlcNAc uptake (30, 49, 51). NgcEFG also internalizes (GlcNAc)2. S. coelicolor A3(2) transports GlcNAc via the PTS enzyme IIC NagE2 as a potentially unique uptake system for GlcNAc when this nutrient is provided as the main carbon source (24), while the uptake of (GlcNAc)2 is mediated by the ABC transporter DasABC (31) for subsequent hydrolysis into GlcNAc by the N-acetyl-β-D-glucosaminidase DasD (33). The catabolism of GlcNAc further requires the GlcNAc kinase NagK, GlcNAc- 6-phosphate deacetylase NagA, and GlcN-6-P deaminase/ isomerase NagB in order to generate fructose-6-phosphate, which will enter glycolysis (39). The expression of all pts, nag, and das genes encoding GlcNAc and (GlcNAc)2 transporters and catabolic enzymes is inhibited by the GntR family transcription factor DasR, the DNA-binding activity of which is repressed by GlcNAc-6-P and GlcN-6-P (6, 9, 22, 23, 27, 41, 43). The expression of all of these genes is activated by GlcNAc, except for the dasA gene, the transcription of which is induced by chitin and (GlcNAc)2 and repressed by GlcNAc (6, 31), similar to the genes encoding chitinase (chi) (20). DasR is required for the maximal expression of chi genes (22), while in the closely related actinobacterium Saccharopolyspora erythraea, DasR acts as a transcriptional repressor of chi genes, similar to other chitin/GlcNAc utilization genes (17).
The DasABC system uses the multiple sugar import protein MsiK as an ATPase (32). The inactivation of msiK abolishes (GlcNAc)2 consumption, whereas the dasA-null mutant maintains the ability to consume (GlcNAc)2, but at a markedly lower rate (31). These findings suggest that there is at least one additional ABC transporter for the uptake of (GlcNAc)2, which also involves MsiK as a common ATPase component (32). In S. coelicolor, the MsiK-mediated uptake of (GlcNAc)2 is required not only for the utilization of chitin degradation byproducts, but also to induce chitinase production (32). However, the inactivation of dasA resulted in stronger total chitinase activity by S. coelicolor, which is not consistent with a simple induction model that requires the transport of (GlcNAc)2 to trigger the chitinolytic system (6, 31). This phenotype suggests that the proper induction of chitin utilization genes needs to involve diverse sensory/transporter systems that act synergistically or competitively according to the extracellular concentration pattern of chitin-derived nutrients (6).
In order to improve our understanding of the chitin utilization system in streptomycetes, we investigated the role of the SCO6005–6007 gene cluster of S. coelicolor, which has a homologous gene organization and genomic context to the genes for the high-affinity GlcNAc and (GlcNAc)2 NgcEFG transporter of S. olivaceoviridis (30, 51). However, while gene synteny is conserved, similarities at the amino acid level between SCO6005–6007 gene products and NgcEFG were low for orthologous proteins. In the present study, we investigated how the lack of similarities between these orthologous transporters impacts on the capacity of the S. coelicolor NgcEFG (NgcEFGSco) system to consume and respond to GlcNAc and (GlcNAc)2 using biochemical and reverse-genetic analyses.
Materials and Methods
Bacterial strains, plasmids, and media
S. coelicolor A3(2) strain M145 (14), its dasA- and dasR-null mutants ASC2 and BAP29 (27, 31), and dasR-overexpressing strain carrying the multicopy dasR gene (dasR++) (26) were used. Escherichia coli JM109 (52) and DH5α (42) were used as hosts for gene manipulation. E. coli ET12567 (dam dcm hsdS) (18) was used to prepare plasmids for S. coelicolor transformation in order to avoid the methylation-specific restriction system of the bacterium. E. coli BL21(DE3)pLysS (Novagen, Burlington, MA, USA) was used to overproduce the NgcESco and DasR proteins. The plasmids used in the present study are listed in Table S1. Luria–Bertani (LB) medium (34) was used to culture S. coelicolor; E. coli transformants were grown in LB medium supplemented with 50 μg mL−1 ampicillin or 10 μg mL−1 gentamycin. Minimal medium (MM; 10 mM K2HPO4, 10 mM KH2PO4, 1 mM CaCl2, and 0.5 mM MgCl2 supplemented with 0.1% [v/v] trace element solution) (35) was used to investigate the responses of S. coelicolor cells to various carbon sources. Soya flour—mannitol (SFM) agar medium (14) was used to prepare spores of S. coelicolor strains.
Gene manipulation
Plasmid preparation and restriction enzyme digestion were performed as described by Sambrook & Russell (2001) (34). DNA fragments were ligated using a DNA ligation kit (Takara Bio, Kusatsu, Japan) according to the manufacturer’s instructions.
Production and purification of recombinant NgcESco and NgcE proteins
Two sets of primers (Table S2) were designed to amplify parts of the SCO6005 (ngcESco) gene, which encode the part of the NgcESco protein without the putative signal peptide (29 amino acids from the N terminus). The recombinant NgcESco protein was tagged with an N-terminal 6×His or N-terminal GST using pET16b or pGEX-4T-1 (Table S1). Both recombinant NgcESco proteins were successfully overproduced in a soluble form and purified using Ni-NTA agarose (Qiagen, Hilden, Germany) and Glutathione Sepharose 4B (GE Healthcare, Waukesha, WI, USA), respectively. The recombinant N-terminally His-tagged NgcE protein of S. olivaceoviridis was also produced in E. coli carrying pQEH301 (Table S1) and purified as reported previously (30). The purified His-tagged NgcESco protein was used to prepare anti-NgcESco antiserum, while binding affinities for the sugars of the purified GST-tagged NgcESco protein were assessed as described below following the removal of the GST-tag. The sugar-binding affinity of the purified His-tagged NgcE protein was also analyzed as described below. See the Supplementary Materials and Methods for detailed conditions pertaining to protein production and purification.
Isothermal titration calorimetry (ITC)
ITC experiments were performed with an iTC200 system (GE Healthcare) (50). Solutions were thoroughly degassed prior to experiments in order to avoid air bubbles in the calorimeter. A volume of 0.2028 mL of NgcESco solution (19 μM) in 20 mM Tris/HCl buffer (pH 8.0) at 30°C was placed in the reaction cell, and ligand solutions in identical buffers were placed in the ITC syringe. In all titrations, 0.8-μL aliquots were injected into the reaction cell at 80-s intervals with a stirring speed of 1,000 rpm. Titrations were completed after 40 injections. The shape of the ITC binding curve was assessed by the Wiseman c value. When titration experiments were performed with c values from 10 to 100 (c=N·Ka·[M]t; where N is stoichiometry, Ka is the association constant, and [M]t is the initial protein concentration), the Ka values obtained were regarded as being reliable (50). ITC data were collected and fit automatically using microcal origin v.7.0 software accompanying the iTC200 system (50). All data from the binding reactions fit well with the single-site binding model yielding stoichiometry (N), an equilibrium dissociation constant (Kd), and enthalpy change (ΔH). The reaction free energy change (ΔG) and entropy change (ΔS) were calculated from the relationship described in the following equation: ΔG=−RTlnKassoc=ΔH−TΔS.
Assessment of binding affinities for sugars based on alterations in fluorescent strength
The Kd value of NgcESco or NgcE was measured against N-acetylglucosamine, N-acetylgalactosamine, N-acetylmuramic acid, glucose, xylose, or mannose based on a fluorescence method (10).
Disruption of the ngcESco gene
The ngcESco gene was disrupted in the wild-type strain S. coelicolor A3(2) M145 and its dasA-null mutant ASC2 (31) by homologous recombination using the temperature-sensitive plasmid pAS100 (Table S1) (51). Most of the ngcESco gene was replaced by the aacC4 gene cassette (Fig. S3 and S4) (3). Detailed methods are described in the Supplementary Materials and Methods.
Complementation of the ngcEFGSco gene cluster
As derivatives of the multi-copy plasmid vector pWHM3 (Table S1) (45), the plasmids pWHM3-ngcEFG and pWHM3-ngcFG were prepared to express ngcEFGSco and ngcFGSco, respectively, with the native promoter region (Fig. S3). Details for constructing these plasmids are provided in the Supplementary Materials and Methods. These constructs were introduced into S. coelicolor strains via protoplast transformation (14).
Conditions for the S. coelicolor culture
In order to investigate the responses of cells to various sugars, we cultured S. coelicolor strains according to a previously described method (31). Spores formed on SFM agar medium were inoculated into 30 mL of LB medium in a 100-mL flask with a spring (14) and grown at 30°C for 18–20 h on a rotary shaker at 150 rpm. Mycelia were harvested by centrifugation (3,000 rpm, 3 min), washed with MM without carbon sources, suspended in 60 mL of MM, and divided into several aliquots. Each aliquot was supplemented with a different carbon source: 250 μM of glucose, maltose, cellobiose, xylobiose, glucosamine, GlcNAc, or (GlcNAc)2 and 0.05% (w/v) colloidal chitin. After sugar supplementation, cultures were again grown at 30°C on a rotary shaker at 150 rpm. In measurements of GlcNAc and (GlcNAc)2 consumption rates, the amount of mycelia in MM was adjusted to 19–21 mg fresh weight mL culture−1. Culture fluids were sampled periodically, centrifuged to separate the supernatant and mycelia, and stored at −80°C. The sugar concentrations and chitinase activities of the supernatants were measured, whereas mycelia were used for total RNA preparation and immunoblot analyses.
Measurement of sugar concentrations
GlcNAc and (GlcNAc)2 concentrations were measured in culture supernatants using high-performance liquid chromatography with UV detection at 215 nm (SPD-20A; Shimadzu, Kyoto, Japan) and a normal phase column of 4.6 mm×250 mm (Inertsil NH2 3 μm; GL Science, Tokyo, Japan). GlcNAc and (GlcNAc)2 were separated under isocratic conditions (acetonitrile/water=65/35 [v/v]) at a flow rate of 1.0 mL min−1 and identified by their respective retention times.
Chitinase assay
Chitinase activity was measured using the fluorescent substrate 4-methylumbelliferyl-N,N′-diacetylchitobioside (Sigma, St. Louis, MO, USA) according to a previously described method (19). One unit of chitinase activity was defined as the amount of enzyme that liberated 1 μmol of 4-methylumbelliferone from the substrate at 37°C in one minute.
Electromobility gel shift assays (EMSAs)
EMSAs were performed using Cy5-labeled dre probes (final concentration, ~0.1 mM) and DasR-6His (final concentration, ~1 mM) in a total reaction volume of 50 μL. The protocol for DasR-6His production from pFT240 (Table S1) (26) in E. coli BL21(DE3) and subsequent purification onto a Ni2+-nitrilotriacetic acid-agarose column was applied as previously described (43). Probes were separated by gel electrophoresis in a 1% (w/v) agarose gel and the fluorescence of the probes was visualized using a Typhoon Trio + variable mode imager (GE Healthcare). The sequences of the oligonucleotides used to generate Cy5-fluorescent double-stranded DNA probes (drenagKA, dredasA, and drenagB) are described in Table S2.
ChIP-on-chip and microarray analysis
ChIP-on-chip and microarray analyses of the DasR binding event on the ngcESco upstream region and the transcription profiles of ngcESco, respectively, were retrieved from raw data published as supplementary files from Świątek-Połatyńska et al. (2015) (41).
Reverse transcription-PCR
DNA-free total RNA was prepared from mycelia using our method (31) and an SV Total RNA Isolation System (Promega, Madison, WI, USA). In order to characterize transcripts, a reverse transcription (RT)-PCR analysis was performed using AccuPower RT/PCR Premix (Bioneer, Daejeon, Korea) as reported previously (31). A set of primers specific for the ngcESco transcript was designed to give a PCR product of 540 bp (Table S2). In PCR, the number of cycles was set to 20 in order to avoid the saturation of PCR product formation. RT-PCR experiments without prior RT were performed in order to ensure that no residual DNA was present in the RNA samples.
In expression studies on dasA, nagE2, and ngcESco in S. coelicolor M145, the RNAs of the dasR null mutant (BAP29) and the strain overexpressing dasR (dasR++) were collected after 30 h of growth in MM mannitol (0.5% [w/v]) agar plates with or with 1% GlcNAc. In the semi-quantitative analysis, samples were taken at four-cycle intervals between cycles 27–35 in order to compare non-saturated PCR product formation (amplifications at cycles 27 and 31 are presented in the first and second wells of each assay). RT-PCR without reverse transcription was performed as a control in order to confirm the absence of residual DNA. Data were verified by three independent experiments.
Immunoblot analysis
S. coelicolor mycelia, which were incubated for 4 h in MM supplemented with 250 μM of each carbon, were harvested by centrifugation (18,000×g, 4°C, 3 min), suspended in phosphate-buffered saline (34), and disrupted by sonication (15 s×8) on ice. The suspension was centrifuged at 10,000×g at 4°C for 5 min, and the protein concentration of the supernatant was measured by Bradford’s method (4). Proteins corresponding to 50 μg were separated with 10% polyacrylamide gels containing 0.1% sodium dodecyl sulfate (16) and blotted onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore, Burlington, MA, USA). Anti-DasA antiserum (31) and anti-NgcESco antiserum, which were prepared using the His-tagged NgcESco protein as an antigen, were used in the immunoblot analysis.
Results
In silico analysis of SCO6005–6007 of S. coelicolor
SCO6005 encodes a putative extracellular sugar-binding component of the transporter (pfam01547), the orthologous protein of which in S. lividans is exported via the twin-arginine translocation (TAT) pathway (11). The gene cluster includes two additional ORFs encoding the putative ABC-type integral membrane proteins (SCO6006 and SCO6007) that form a transporter permease (Fig. S1). Regarding most streptomycetes sugar ABC transporters, the gene for the ATPase component was not included in the cluster and energy for sugar import was most likely provided by the multiple sugar import ATPase MsiK (13, 32, 36, 37, 46). The Rok family regulatory gene rok7B7 is immediately downstream of the operon, and controls the xylose operon SCO6009–6011 (40). Upstream of SCO6005, 6004 encodes a putative alpha-1,2-mannosidase.
The SCO6005–6007 operon of S. coelicolor is an orthologue of the S. olivaceoviridis ngcEFG operon, which encodes a high-affinity transporter for GlcNAc and (GlcNAc)2 (30, 51). While gene synteny is strictly conserved in streptomycetes, identity at the amino acid level between SCO6005–6007 gene products and NgcEFG is low for orthologous proteins, namely 35% amino acid identity for the SCO6005 protein and NgcE, 44% for SCO6006 and NgcF, and 50% between SCO6007 and NgcG (Fig. S1). In contrast, the other streptomycetes NgcE orthologues share between 80 to 91% amino acid identities throughout the full-length sequence. These low amino acid identities between S. coelicolor and S. olivaceoviridis and other streptomycetes are limited to the three Ngc proteins because the putative products of adjacent ORFs SCO6004 and SCO6008 (ROK7B7) present high levels of identity, as expected for orthologous proteins.
Binding specificity of the NgcESco protein
The lack of identity between Ngc proteins from S. coelicolor and S. olivaceoviridis prompted us to assess the binding affinity of the solute-binding component of the transporter of S. coelicolor (NgcESco). The binding specificity and affinity of the pure NgcESco protein heterologously produced in E. coli (see Materials and Methods for details) was initially investigated using ITC. As shown in Fig. S2, the quantity of heat of the NgcESco solution increased with the concentrations of GlcNAc and (GlcNAc)2, but was not affected by the addition of (GlcNAc)3 and higher oligomers up to (GlcNAc)6, thereby demonstrating that the recombinant NgcESco protein interacted with GlcNAc and (GlcNAc)2. NgcESco and GlcNAc/(GlcNAc)2 bound in a 1:1 stoichiometry and binding in both cases was driven by enthalpy, while the loss of entropy opposed binding, suggesting a specific interaction between NgcESco and GlcNAc/ (GlcNAc)2 (Table 1). Kd values for GlcNAc and (GlcNAc)2 were 1.15 and 1.53 μM, respectively (Table 1). These values were higher than those of S. olivaceoviridis NgcE for GlcNAc and (GlcNAc)2 i.e., 8.3 and 29 nM, respectively (51), and that of DasA for (GlcNAc)2 i.e., 32 nM (31).
Table 1
Thermodynamic parameters for
N-acetylglucosamine (GlcNAc) and
N,N′-diacetylchitobiose ([GlcNAc]
2) binding to NgcE
Sco obtained from ITC profiles shown in Fig. S2.
| Ligand |
N |
Kd (μM) |
ΔH (kcal mol−1) |
ΔS (cal mol−1 K−1) |
−TΔS (kcal mol−1) |
ΔG (kcal mol−1) |
| GlcNAc |
1.05 |
1.15 |
−9.98 |
−5.77 |
1.75 |
−8.23 |
| (GlcNAc)2 |
0.98 |
1.53 |
−12.3 |
−14.0 |
4.24 |
−8.06 |
N, binding stoichiometry; Kd, dissociation constant; ΔH, change in enthalpy; ΔS, change in entropy; T, temperature; ΔG, change in Gibbs free energy.
In order to more precisely compare the affinity of NgcESco with that of NgcE, the recombinant NgcESco and NgcE proteins produced in E. coli were purified and their affinities were evaluated based on changes in the fluorescent strengths of the proteins. The addition of GlcNAc did not quench the fluorescent strengths of the proteins, it increased them. Kd values were calculated based on increments in the fluorescent strength after the addition an increasing amount of GlcNAc. The Kd value of NgcESco for GlcNAc was 1.9 μM, which corresponded with that obtained by ITC (Table 1). The Kd value of NgcE produced in E. coli for GlcNAc was 85 nM. Although this value was one magnitude higher than that obtained by surface plasmon resonance, it was still 22-fold lower than that of NgcESco, indicating the markedly higher affinity of the NgcE protein. (GlcNAc)2 did not modify the fluorescence properties of NgcESco or NgcE. The Kd values of NgcESco for N-acetylgalactosamine (GalNAc) and N-acetylmuramic acid (MurNAc) were 12 and 25 μM, respectively, and were 6- and 13-fold higher than that for GlcNAc (1.9 μM). We also investigated the effects of xylose and mannose on the fluorescent strength of NgcESco due to the presence of genes coding for putative mannosidase and a regulator of the xylose operon in the vicinity of the ngcEFG operon (Fig. S1). Glucose, xylose, or mannose up to 1 mM did not significantly affect the fluorescent strength of NgcESco, implying the absence of an interaction between NgcESco and these sugars. The Kd values of the maltose-binding protein (MBP), L-arabinose-binding protein (ABP), and D-glucose/ D-galactose-binding protein (GGBP) of ABC transporters for the corresponding ligand sugars range between 10−8 and 10−6 M (25). The Kd values of NgcESco for GlcNAc and (GlcNAc)2 were in the 10−6 M range (Table 1), implying that the protein mediates the uptake of these sugars; however, affinities were lower than those of S. olivaceoviridis NgcE for GlcNAc and (GlcNAc)2.
(GlcNAc)2 and GlcNAc consumption in the ngcESco mutant
The ngcESco gene was disrupted in S. coelicolor strain M145 and its dasA null-mutant ASC2 (Fig. S3 and S4) in order to assess its contribution to GlcNAc and/or (GlcNAc)2 uptake. The mycelia of strains M145, ASC2, the ngcESco-null mutant (strain CI1), and dasA-ngcESco double-null mutant (strain CI3), pregrown in LB medium, were cultivated in MM supplemented with 250 μM of GlcNAc or (GlcNAc)2. GlcNAc consumption rates were not significantly affected by the disruption of ngcESco regardless of whether they were examined in the wild-type- or dasA-minus background (Fig. 1A). The disruption of msiK lowered the rate of GlcNAc consumption (Fig. 1A), suggesting the presence of ABC transporter(s) for GlcNAc uptake.

In contrast to GlcNAc, the (GlcNAc)2 consumption pattern was affected by the ngcESco mutation (Fig. 1B). (GlcNAc)2 consumption in the wild-type strain M145 was divided into two steps: the initial step from 0–2 h (2.3 nmol h−1 mg mycelia−1 [R2=0.991]) and the next induced step from 2–3 h (6.0 nmol h−1 mg mycelia−1 [R2=0.998]). In the ngcESco-null mutant, (GlcNAc)2 consumption was delayed (Fig. 1B). In the first step during 0–2 h, the (GlcNAc)2 concentration remained almost constant, and the initiation of the next induced consumption was delayed for 0.5–1 h. The consumption rate of induced consumption (3–4 h) in CI1 was 8.6 nmol h−1 mg mycelia−1 (R2=0.998). The dasA-null mutant ASC2 consumed (GlcNAc)2 constantly (2.8 nmol h−1 mg mycelia−1 [0–5 h, R2=0.993]), as reported previously (31).
In the dasA-minus background, the effects of the disruption of ngcESco were more obvious. The dasA-ngcESco double mutant CI3 showed a low level of (GlcNAc)2 consumption (1.2 nmol h−1 mg mycelia−1 [2–5 h, R2=0.986]). The msiK-null mutant ASC3 had the lowest consumption rate (0.6 nmol h−1 mg mycelia−1 [2–7 h, R2=0.960]) among the strains tested. These results indicate that the ngcESco gene is involved in (GlcNAc)2 uptake in S. coelicolor M145, particularly in the initial and constant consumption prior to the induction of the DasABC-MsiK transporter.
Chitinase production in the ngcESco mutant
We previously reported that (GlcNAc)2 uptake is necessary for the induction of chitinase production in S. coelicolor (32). In order to elucidate the involvement of NgcESco in chitinase production, the effects of the disruption of ngcESco on chitinase production were investigated. As shown in Fig. 2A, the chromosomal deletion of ngcESco reduced the level of chitinase activity induced in the presence of (GlcNAc)2. In contrast, the dasA-ngcESco double mutation fully abolished chitinase production in the presence of (GlcNAc)2 (Fig. 2A), as observed for the msiK-null mutant ASC3 (32). The dasA mutant, which had a lower (GlcNAc)2 consumption rate than M145 (Fig. 1A), exhibited stronger chitinase activity (Fig. 2A), as reported previously (31). The delay in chitinase production (Fig. 2B) was reproducibly observed in CI1 when colloidal chitin was added, suggesting that NgcE is involved in sensing chitin and triggering the chitinolytic system. The double mutant CI3 showed partial chitinolytic activity in the presence of colloidal chitin after a prolonged incubation (8–10 d) (Fig. 2B). Complementation experiments revealed that the dasA-ngcESco double mutant CI3 recovered the induction of chitinase production by introducing a multi-copy plasmid carrying ngcEFGSco with its native promoter, whereas it did not with a plasmid only carrying ngcFSco and ngcGSco encoding the membrane component of the transporter (Fig. 2C and S3). Similar to the dasA mutant (Fig. 2A), the induced level of chitinase activity was markedly higher in strain CI3, which carries the ngcEFGSco operon on a multi-copy plasmid (pWHM3-ngcEFG), than in CI1, which is the ngcESco mutant carrying the empty vector (pWHM3) (Fig. 2C). The production of NgcESco in complemented strain CI3 (pWHM3-ngcEFG) was confirmed by the immunoblot analysis using anti-NgcESco antiserum (Fig. S5).

In order to elucidate the roles of the distinctive transporters in the induction of the chitinolytic system, we assessed chitinase production profiles in the presence of lower concentrations of (GlcNAc)2. At 50 μM of (GlcNAc)2, the dasA mutant exhibited stronger chitinolytic activities than the parental strain M145 and its ngcESco mutant CI1 (Fig. 2D). At 5 μM of (GlcNAc)2, the level of chitinase activity in the dasA mutant was similar to that in the presence of 50 μM (GlcNAc)2 in M145 and CI3, while the ngcESco mutant and the parental strain M145 exhibited very weak chitinase activities at this concentration (Fig. 2D).
Regulation of ngcESco expression
A ChIP-on-chip approach for S. coelicolor M145 carrying the integrative vector pGAM29, which expresses C-terminally 3×FLAG-tagged DasR (see [41] for details), revealed DasR binding to the intergenic region between SCO6004 and SCO6005 (ngcESco) (Fig. 3A). This region possesses the predicted DasR responsive element (dre) AGTGGACTATACCTGT at nt position −334 upstream of SCO6005 (drengcE) (Fig. 3A), which matches 12 out of the 16 nt of the dre consensus sequence (5). The DasR-binding event was abolished when S. coelicolor was grown in the presence of GlcNAc (Fig. 3A). In order to confirm ChIP-on-chip data, EMSAs were performed using pure His-tagged DasR (DasR-6His) and a short double-stranded oligonucleotide centered on drengcE (Table S2). DasR interacted with the DNA probe containing drengcE, as observed with the positive control probes containing dre upstream of nagKA and dasA (drenagKA and dredasA) (Fig. 3B). The binding of DasR to the drengcE-containing probe was inhibited by GlcNAc-6P and GlcN-6P (Fig. 3C). GlcNAc-6P inhibited binding more efficiently than GlcN-6P (Fig. 3C). These results of the ChIP-on-chip analysis and EMSAs were consistent with those reported for the interactions of DasR with other dre (41, 43). GlcNAc-derived GlcNAc-6P and GlcN-6P inhibited the binding of DasR to dre in the ChIP-on-chip analysis.
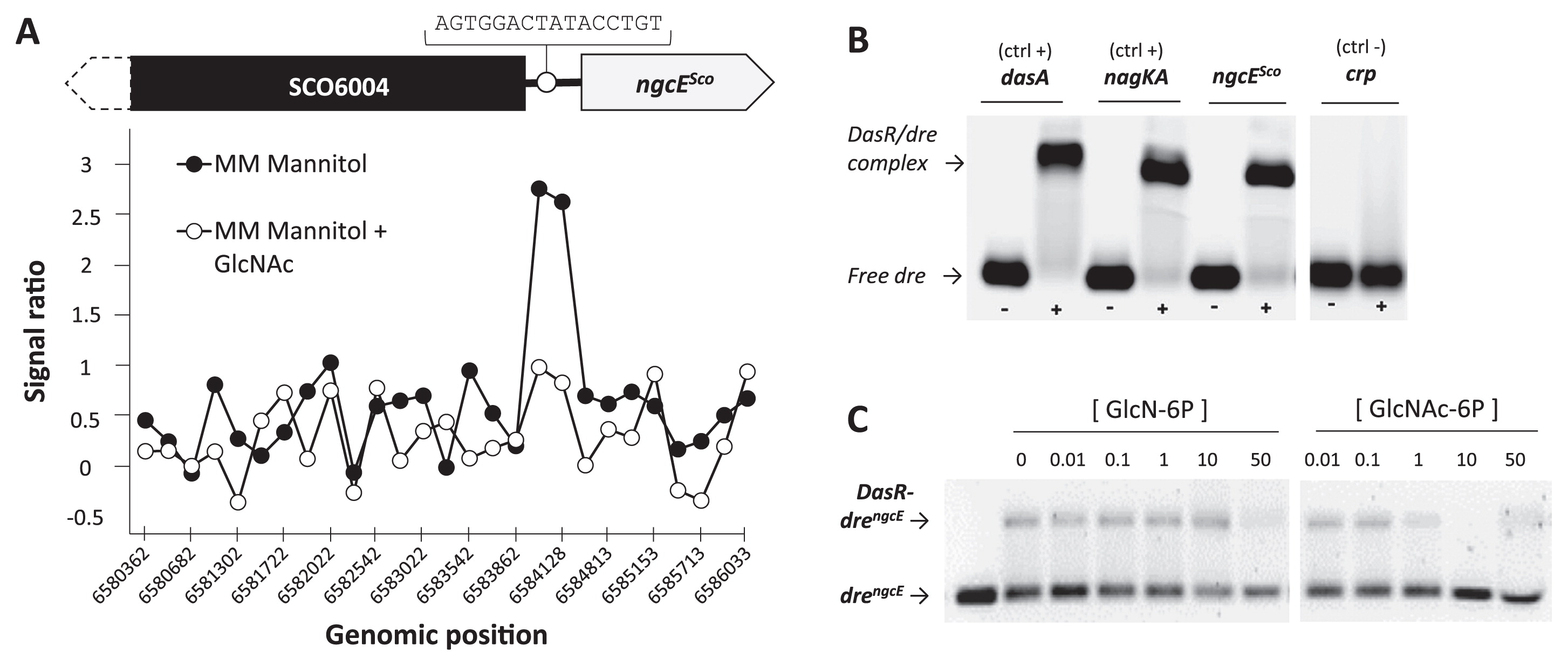
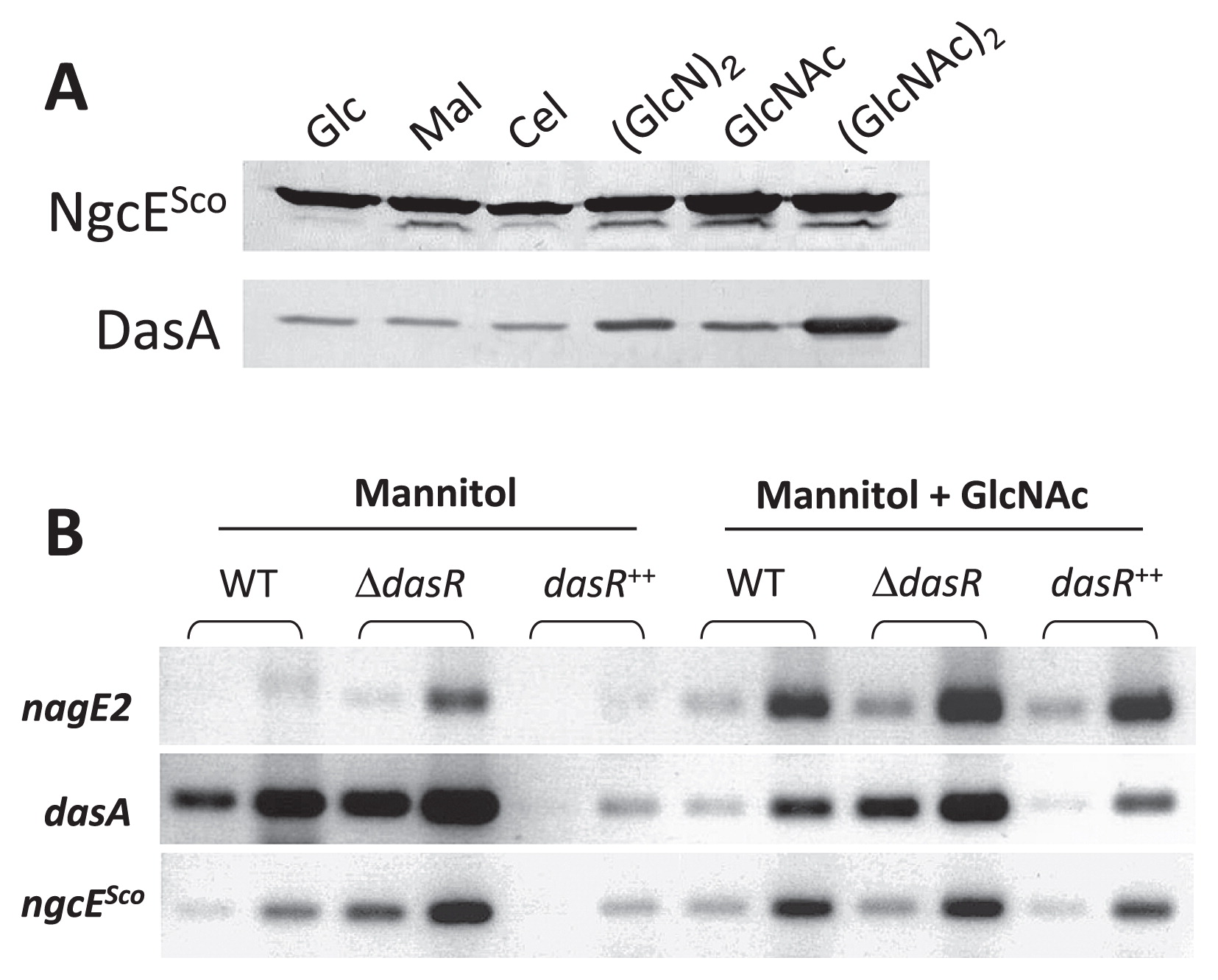
Previous transcriptomic studies also revealed that ngcESco expression was induced by chitin (23) and GlcNAc (41). The microarray analysis revealed that the expression of ngcESco was up-regulated in the dasR mutant in the absence of GlcNAc and appeared to be induced at earlier time points (24 and 30 h) when S. coelicolor M145 was grown in MM medium supplied with GlcNAc (Fig. S6A). Very similar expression profiles were observed for ngcFSco and ngcGSco (41), suggesting that ngcESco (SCO6005), ngcFSco (SCO6006), and ngcGSco (SCO6007) form a tri-cistronic operon that was herein confirmed using the RT-PCR analysis (Fig. S6B). In the dasR mutant, ngcESco transcription was not induced by GlcNAc (Fig. S6A). When mycelia grown in LB were exposed to 250 μM glucose, maltose, cellobiose, xylobiose, GlcNAc, or (GlcNAc)2, the amounts of ngcESco transcripts were similar among the tested conditions (Fig. S6C), whereas dasA transcription was strongly induced in the presence of (GlcNAc)2 under the same culture conditions (31, 32). In order to investigate the expression of ngcESco at the level of protein production, an immunoblot analysis was performed using antibodies against the recombinant His-tagged NgcESco protein overproduced in E. coli. NgcESco production was observed in the presence of glucose, maltose, cellobiose, (GlcN)2, GlcNAc, or (GlcNAc)2 (Fig. 4A). The levels of production in the presence of GlcNAc and (GlcNAc)2 were 1.3- and 1.4-fold higher than that in the presence of glucose, respectively. In contrast, DasA production was markedly induced by (GlcNAc)2 and by the glucosamine dimer (GlcN)2, though to a markedly lower degree (Fig. 4A). Since the abundant carbon and nitrogen sources contained in LB medium may affect ngcESco transcriptional responses to amino sugars, we repeated the expression studies on RNA samples that were prepared from mycelia grown on MM mannitol (0.5% [w/v]) with or without GlcNAc (1.0% [w/v]) at 28°C for 30 h. Under these conditions, the transcription of ngcESco was stronger in the dasR mutant and weaker in the dasR++ strain than in the parental strain M145, demonstrating that DasR acts as a transcriptional repressor of ngcESco under these conditions (Fig. 4B). Similar to that observed for nagE2, the transcription of ngcESco was induced when GlcNAc was added to MM mannitol in the wild-type or dasR++ strain. However, in the dasR mutant, ngcESco transcription was not further enhanced in the presence of GlcNAc, as previously observed in a transcriptomic analysis (Fig. S6A) (41).
Discussion
In the present study, we investigated the role of the ngcESco gene (SCO6005) and its encoding protein NgcESco in order to assess its contribution to the uptake and catabolism of chitin and its main byproducts GlcNAc and (GlcNAc)2. As discussed in the Introduction, we were unable to strictly refer to a previous study performed on ngcE in S. olivaceoviridis because despite the conserved synteny, the level of identity with NgcESco was only 35% (Fig. S1). The lack of amino acid identity between the two orthologues is reflected in the Kd values of NgcESco measured for GlcNAc and (GlcNAc)2 (1.15 and 1.53 μM, respectively [Table 1]), which were higher than those of the S. olivaceoviridis NgcE protein for GlcNAc and (GlcNAc)2 (8.3 and 29 nM, respectively) (51), and the Kd value of DasA for (GlcNAc)2 (32 nM) (31). The expression of ngcESco was constitutive and induced to some extent by GlcNAc and (GlcNAc)2, while dasA expression was leaky and strongly induced by (GlcNAc)2 (Fig. 4B and S6B). The initial (GlcNAc)2 consumption rate in M145 (2.3 nmol h−1 mg mycelia−1) corresponded well with the constant (GlcNAc)2 consumption rate (2.8 nmol h−1 mg mycelia−1) in its dasA mutant, whereas the dasA-ngcESco and msiK mutants had markedly lower rates (1.2 and 0.6 nmol h−1 mg mycelia−1, respectively) (Fig. 1B). Therefore, we suggest that NgcESco acts as the constitutive sugar-binding protein of the ABC transporter NgcEFGSco-MsiK for the uptake of (GlcNAc)2 in S. coelicolor A3(2), while DasABC-MsiK is the main (GlcNAc)2 uptake system, the production of which is strongly induced by (GlcNAc)2. When consumption experiments were performed with various amounts of mycelia (5–15 mg mycelia mL−1), the effects of the ngcESco mutation on (GlcNAc)2 consumption were negligible, in contrast to the disruption of dasA, which markedly reduced the (GlcNAc)2 consumption rate (data not shown), possibly reflecting the 50-fold higher Kd value of NgcESco for (GlcNAc)2 than that of DasA. We assumed that remaining (GlcNAc)2 consumption in the dasA-ngcESco and msiK mutants was due to (GlcNAc)2 hydrolysis based on the basal level of extracellular N-acetylhexosaminidases and subsequent consumption of GlcNAc.
The reverse-genetic analysis did not indicate the involvement of ngcESco in the uptake of GlcNAc. The NgcEFGSco- MsiK system may not uptake GlcNAc even though NgcESco interacts with GlcNAc. The MalE protein, which is the maltose (maltodextrin)-binding protein for the uptake of maltose and maltodextrin in E. coli, interacts with ligands and mediates the uptake of sugars. Reduced or oxidized maltodextrins were not transported into cells, but bound to MalE with good affinity (8). Similarly, the “maltodextrin-negative” mutants of MalE only show a marginal decrease in affinity toward maltodextrins, but do not support the transport of maltodextrins in whole cells (48).
In the present study, we observed a reduced GlcNAc consumption rate in ASC3 (Fig. 1A) that lacks the msiK gene encoding the common ATPase component for sugar ABC transporters (32). These results imply the presence of ABC transporters for GlcNAc; however, a previous study reported that the NagE2 of PTS may be a unique permease mediating the uptake of GlcNAc in S. coelicolor (24).
The presence of higher (DasABC) and lower (NgcEFGSco) affinity uptake systems for (GlcNAc)2 in S. coelicolor is likely to have a biological meaning. Similarly, in S. olivaceoviridis, the uptake of GlcNAc is mediated by two systems, the affinities of which are distinctive: the Km value of one system (the PTS system including PtsC2) for 14C-labeled GlcNAc is 5 μM, while that of the other system (ABC transporter containing NgcEFG) is 0.48 μM (30, 49).
The ngcESco-dasA double mutation abolished the induction of chitinase production by (GlcNAc)2 as the msiK mutation (Fig. 2A). These results clearly demonstrated that the uptake of (GlcNAc)2 is essential for the induction of chitinase production, as concluded in our previous study (32). It was noteworthy that the single ngcESco and dasA mutants exerted contrasting effects on the induction of chitinase production. The disruption of ngcESco reduced the chitinase activity induced by (GlcNAc)2, while the dasA mutation increased not only the levels of induced chitinase activity in the presence of (GlcNAc)2 or colloidal chitin, but also sensitivity to (GlcNAc)2 (Fig. 2A, B, and D). This result implies distinct roles for the two (GlcNAc)2 transporters. We assume that DasABC acts in the metabolism of (GlcNAc)2. The structures of the ngcEFGSco and dasABC gene clusters imply roles for the encoding ABC transporters for (GlcNAc)2 uptake; a gene for the N-acetylglucosaminidase DasD hydrolyzing (GlcNAc)2 to GlcNAc is present in the dasABC gene cluster, whereas such a gene involved in (GlcNAc)2 hydrolysis is not clustered with ngcEFGSco (Fig. S1). The disruption of dasD increased the level of chitinase production in the presence of (GlcNAc)2 or chitin (33). The dasD mutation may prolong the life of intracellular (GlcNAc)2, which induces chitinase production (33). We assumed that the higher sensitivity of the dasA mutant to (GlcNAc)2 in chitinase production (Fig. 2D) is attributed to the longer life of intracellular (GlcNAc)2, which induces the expression of chi genes. In contrast, the reduction in chitinase activity induced by (GlcNAc)2 in the ngcESco mutant may be ascribed to the shorter life of the disaccharide.
NgcESco did not appear to be essential for the uptake of (GlcNAc)2 or induction of chitinase production (Fig. 1B, 2A, and B). However, it is involved in these processes and may have roles in the initial accumulation of intracellular (GlcNAc)2 for sensing chitin as a nutrient source in the environment. This hypothesis is supported by the observed late induction of chitinase production in the presence of colloidal chitin and the low initial (GlcNAc)2 consumption rate in the ngcESco mutant (Fig. 1B and 2B). In the presence of chitin, (GlcNAc)2 is expected to be continuously generated by chitin hydrolysis with extracellular chitinases produced leakily (or possibly by the chitinases of other microorganisms in ecosystems), and continually taken up mainly via NgcEFGSco-MsiK (Fig. 4B) until the (GlcNAc)2 concentration becomes sufficient to trigger the expression of das and chi (Fig. 5). Therefore, the intracellular accumulation of (GlcNAc)2 and subsequent induction of chitinase production may be delayed in the ngcESco mutant in the presence of colloidal chitin.

In the ngcESco-dasA mutant, the induction of chitinase production by colloidal chitin was markedly delayed (Fig. 2B). Chitinase production in the presence of colloidal chitin was abolished in the msiK mutant (Fig. 2B) (32), which implies the presence of additional ABC transporters for (GlcNAc)2 or the heterologous disaccharide GlcNAc-GlcN and/or GlcN-GlcNAc, which may be produced by the hydrolysis of colloidal chitin.
The results of RT-PCR, immunoblot assays, and previous transcriptomic and ChIP-on-chip analyses indicate that the expression of ngcESco is repressed by DasR and induced by GlcNAc, (GlcNAc)2, and chitin (22), though with a markedly weaker induction response to these elicitors than dasA in the presence of (GlcNAc)2 and nagE2 by GlcNAc (Fig. 3 and 4). It is noteworthy that the control of ngcESco expression is unique because it is the only known DasR-controlled gene that is induced by GlcNAc, (GlcNAc)2, and chitin. The in vivo binding pattern of DasR to drengcE differed from the patterns of the genes for DasA, chitinases, and GlcNAc metabolism. DasR binding to drengcE was inhibited by the presence of GlcNAc in MM, whereas DasR bound to the dre of dasA and chitinase genes (chiA, C, D, H, I, and J) (41). In R5 (nutrient rich) medium, DasR binding to the dre of the GlcNAc metabolic genes nagE2 and nagKA was inhibited in the presence of GlcNAc, whereas DasR remained bound to drengcE (41). Although we concluded that NgcESco acted as a component of the ABC transporter for (GlcNAc)2 in the present study, other physiological roles need to be investigated and elucidated.
Acknowledgements
The authors thank Yusuke Kimura at SIST, Tomonori Shinya, Yoshitake Desaki, and Naoto Shibuya at Meiji University, and Tamo Fukamizo at Kinki University for their help with experiments. This work was supported in part by KAKENHI grants 17780056 (2005–2007), 21780064 (2009–2010), and 25292040 (2013–2015) from the Ministry of Education, Science, Sports, and Culture and by the 2008 Young Investigator Research Grant to AS from the Noda Institute for Scientific Research. ET’s work was supported by a FRIA grant and by the Belgian program of Interuniversity Attraction Poles initiated by the Federal Office for Scientific Technical and Cultural Affairs (PAI no. P7/44) and by the FNRS (research project RFNRS.3342-T.0006.14-PDR [FRFC]). SR is an FRS-FNRS research associate.
References
- 1. Barka, E.A., P. Vatsa, L. Sanchez, N. Gavaut-Vaillant, C. Jacquard, H.P. Klenk, C. Clément, Y. Oudouch, and G.P. van Wezel. 2016. Taxonomy, physiology, and natural products of the Actinobacteria. Microbiol Mol Biol Rev. 80:1-43.
- 2. Bérdy, J. 2005. Bioactive microbial metabolites. J Antibiot (Tokyo). 58:1-26.
- 3. Blondelet-Rouault, M.H., J. Weiser, A. Lebrihi, P. Branny, and J.L. Pernodet. 1997. Antibiotic resistance gene cassettes derived from the omega interposon for use in E. coli and Streptomyces. Gene. 190:315-317.
- 4. Bradford, M.M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 72:248-254.
- 5. Colson, S., J. Stephan, T. Hertrich, A. Saito, G.P. van Wezel, F. Titgemeyer, and S. Rigali. 2007. Conserved cis-acting elements upstream of genes composing the chitinolytic system of streptomycetes are DasR-responsive elements. J Mol Microbiol Biotechnol. 12:60-66.
- 6. Colson, S., G.P. van Wezel, M. Craig, E.E.E. Noens, H. Nothaft, A.M. Mommaas, F. Titgemeyer, B. Joris, and S. Rigali. 2008. The chitobiose-binding protein, DasA, acts as a link between chitin utilization and morphogenesis in Streptomyces coelicolor A3(2). Microbiology. 154:373-382.
- 7. Derouaux, A., D. Dehareng, E. Lecocq, et al. 2014. Crp of Streptomyces coelicolor is the third transcription factor for the large CRP-FNR superfamily able to bind cAMP. Biochem Biophys Res Commun. 325:983-990.
- 8. Ferenci, T., M. Muir, K.-S. Lee, and D. Maris. 1986. Substrate specificity of the Escherichia coli maltodextrin transport system and its component proteins. Biochim Biophys Acta. 860:44-50.
- 9. Fillenberg, S.B., M.D. Friess, S. Körner, R.A. Böckmann, and Y.A. Muller. 2016. Crystal structures of the global regulator DasR from Streptomyces coelicolor: implications for the allosteric regulation of GntR/HutC repressors. PLoS One. 11:e0157691.
- 10. Fukamizo, T., S. Amano, K. Yamaguchi, et al. 2005. Bacillus circulans MH-K1 chitosanase: amino acid residues responsible for substrate binding. J Biochem. 138:563-569.
- 11. Guimond, J., and R. Morosoli. 2008. Identification of Streptomyces lividans proteins secreted by the twin-arginine translocation pathway following growth with different carbon sources. Can J Microbiol. 54:549-558.
- 12. Hodgson, D.A. 2000. Primary metabolism and its control in streptomycetes: a most unusual group of bacteria. Adv Microb Physiol. 42:47-238.
- 13. Hurtubise, Y., F. Shareck, D. Kluepfel, and R. Morosoli. 1995. A cellulase/xylanase-negative mutant of Streptomyces lividans 1326 defective in cellobiose and xylobiose uptake is mutated in a gene encoding a protein homologous to ATP-binding proteins. Mol Microbiol. 17:367-377.
- 14. Kieser, T., M.J. Bibb, M.J. Buttner, K.F. Chater, and D.A. Hopwood. 2000. Practical Streptomyces Genetics. John Innes Foundation, Norwich.
- 15. Kubota, T., K. Miyamoto, M. Yasuda, Y. Inamori, and H. Tsujibo. 2004. Molecular characterization of an intracellular β-N- acetylglucosaminidase involved in the chitin degradation system of Streptomyces thermoviolaceus OPC-520. Biosci Biotechnol Biochem. 68:1306-1314.
- 16. Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680-685.
- 17. Liao, C., S. Rigali, C.L. Cassani, E. Marcellin, L.K. Nielsen, and B.C. Ye. 2014. Control of chitin and N-acetylglucosamine utilization in Saccharopolyspora erythraea. Microbiology. 160:1914-1928.
- 18. MacNeil, D.J., K.M. Gewain, C.L. Ruby, G. Dezeny, P.H. Gibbons, and T. MacNeil. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene. 111:61-68.
- 19. Miyashita, K., T. Fujii, and Y. Sawada. 1991. Molecular cloning and characterization of chitinase genes from Streptomyces lividans 66. J Gen Microbiol. 137:2065-2072.
- 20. Miyashita, K., T. Fujii, and A. Saito. 2000. Induction and repression of a Streptomyces lividans chitinase gene promoter in response to various carbon sources. Biosci Biotechnol Biochem. 64:39-43.
- 21. Nakagawa, Y.S., M. Kudo, J.S.M. Loose, T. Ishikawa, K. Totani, V.G. Eijsink, and G. Vaaje-Kolstad. 2015. A small lytic polysaccharide monooxygenase from Streptomyces griseus targeting a- and b-chitin. FEBS J. 282:1065-1079.
- 22. Nazari, B., A. Saito, M. Kobayashi, K. Miyashita, Y. Wang, and T. Fujii. 2012. High expression levels of chitinase genes in Streptomyces coelicolor A3(2) grown in soil. FEMS Microbiol Ecol. 77:623-635.
- 23. Nazari, B., M. Kobayashi, A. Saito, A. Hassaninasab, K. Miyashita, and T. Fujii. 2013. Chitin induces gene expression in secondary metabolic pathways in Streptomyces coelicolor A3(2) grown in soil. Appl Environ Microbiol. 79:707-713.
- 24. Nothaft, H., S. Rigali, B. Boomsma, M. Swiatek, K.J. McDowall, G.P. van Wezel, and F. Titgemayer. 2010. The permease gene nagE2 is the key to N-acetylglucosamine sensing and utilization in Streptomyces coelicolor and is subjected to multi-level control. Mol Microbiol. 75:1133-1144.
- 25. Quiocho, F.A., J.C. Spurlino, and L.E. Rodseth. 1997. Extensive features of tight oligosaccharide binding revealed in high-resolution structures of the maltodextrin transport/chemosensory receptor. Structure. 5:997-1015.
- 26. Rigali, S., M. Schlicht, P. Hoskisson, H. Nothaft, M. Merzbacher, B. Joris, and F. Titgemeyer. 2004. Extending the classification of bacterial transcription factors beyond the helix-turn-helix motif as an alternative approach to discover new cis/trans relationships. Nucleic Acids Res. 32:3418-3426.
- 27. Rigali, S., H. Nothaft, E.E. Noens, et al. 2006. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol Microbiol. 61:1237-1251.
- 28. Saito, A., T. Fujii, and K. Miyashita. 1999. Chitinase system in Streptomyces. Actinomycetologica. 13:1-10.
- 29. Saito, A., G. Biucobić, K. Miyashita, and H. Schrempf. 2001. Characteristics of a Streptomyces coelicolor A3(2) extracellular protein targeting chitin and chitosan. Appl Environ Microbiol. 67:1268-1273.
- 30. Saito, A., and H. Schrempf. 2004. Mutational analysis of the binding affinity and transport activity for N-acetylglucosamine mediated by the novel ABC transporter Ngc within the chitin-degrader Streptomyces olivaceoviridis. Mol Genet Genomics. 271:545-553.
- 31. Saito, A., T. Shinya, K. Miyamoto, et al. 2007. The dasABC gene cluster, adjacent to dasR, encodes a novel ABC transporter for the uptake of N, N′-diacetylchitobiose in Streptomyces coelicolor A3(2). Appl Environ Microbiol. 73:3000-3008.
- 32. Saito, A., T. Fujii, T. Shinya, N. Shibuya, A. Ando, and K. Miyashita. 2008. The msiK gene, encoding the ATP-hydrolyzing component of N,N′-diacetylchitobiose ABC transporters, is essential for induction of chitinase production in Streptomyces coelicolor A3(2). Microbiology. 154:3358-3365.
- 33. Saito, A., H. Ebise, Y. Orihara, et al. 2013. Enzymatic and genetic characterization of the DasD protein possessing N-acetyl-β-d-glucosaminidase activity in Streptomyces coelicolor A3(2). FEMS Microbiol Lett. 340:33-40.
- 34. Sambrook, J., and D.W. Russell. 2001. Molecular Cloning: A Laboratory Manual, 3rd ed. Cold Spring Harbor Laboratory Press, New York.
- 35. Schlochtermeier, A., S. Walter, J. Schröder, M. Moorman, and H. Schrempf. 1992. The gene encoding the cellulase (Avicelase) Cel1 from Streptomyces reticuli and analysis of protein domains. Mol Microbiol. 6:3611-3621.
- 36. Schlösser, A., T. Kampers, and H. Schrempf. 1997. The Streptomyces ATP-binding component MsiK assists in cellobiose and maltose transport. J Bacteriol. 179:2092-2095.
- 37. Schlösser, A. 2000. MsiK-dependent trehalose uptake in Streptomyces reticuli. FEMS Microbiol Lett. 184:187-192.
- 38. Schnellmann, J., A. Zeltins, H. Blaak, and H. Schrempf. 1994. The novel lectin-like protein CHB1 is encoded by a chitin-inducible Streptomyces olivaceoviridis gene and binds specifically to crystalline a-chitin of fungi and other organisms. Mol Microbiol. 13:807-819.
- 39. Świątek, M.A., E. Tenconi, S. Rigali, and G.P. van Wezel. 2012. Functional analysis of the N-acetylglucosamine metabolic genes of Streptomyces coelicolor and role in control of development and antibiotic production. J Bacteriol. 194:1136-1144.
- 40. Świątek, M.A., J. Gubbens, G. Bucca, E. Song, Y.H. Yang, E. Laing, B.G. Kim, C.P. Smith, and G.P. van Wezel. 2013. The ROK family regulator Rok7B7 pleiotropically affects xylose utilization, carbon catabolite repression, and antibiotic production in Streptomyces coelicolor. J Bacteriol. 195:1236-1248.
- 41. Swiatek-Polatynska, M.A., G. Bucca, E. Laing, J. Gubbens, F. Titgemeyer, C.P. Smith, S. Rigali, and G.P. van Wezel. 2015. Genome-wide analysis of in vivo binding of the master regulator DasR in Streptomyces coelicolor identifies novel non-canonical targets. PLoS One. 10e0122479.
- 42. Taylor, R.G., D.C. Walker, and R.R. McInnes. 1993. E. coli host strains significantly affect the quality of small scale plasmid DNA preparations used for sequencing. Nucleic Acids Res. 21:1677-1678.
- 43. Tenconi, E., M. Urem, M.A. Świątek-Połatyńska, F. Titgemeyer, Y.A. Muller, G.P. van Wezel, and S. Rigali. 2015. Multiple allosteric effectors control the affinity of DasR for its target sites. Biochem Biophys Res Commun. 464:324-329.
- 44. Tsujibo, H., N. Hatano, T. Mikami, A. Hirasawa, K. Miyamoto, and Y. Inamori. 1998. A novel β-N-acetylglucosaminidase from Streptomyces thermoviolaceus OPC-520: Gene cloning, expression, and assignment to family 3 of the glycosyl hydrolases. Appl Environ Microbiol. 64:2920-2924.
- 45. Vara, J., M. Lewandowska-Skarbek, Y.G. Wang, S. Donadio, and C.R. Hutchinson. 1989. Cloning of genes governing the deoxysugar portion of the erythromycin biosynthesis pathway in Saccharopolyspora erythraea (Streptomyces erythreus). J Bacteriol. 171:5872-5881.
- 46. Viens, P., M.P. Debeau, A. Kimura, Y. Desaki, T. Shinya, N. Shibuya, A. Saito, and R. Brzezinski. 2015. Uptake of chitosan-derived D-glucosamine oligosaccharides in Streptomyces coelicolor A3(2). FEMS Microbiol Lett. 362fnv048.
- 47. Vrancken, K., and J. Anné. 2009. Secretory production of recombinant proteins by Streptomyces. Future Microbiol. 4:181-188.
- 48. Wandersman, C., M. Schwartz, and T. Ferenci. 1979. Escherichia coli mutants impaired in maltodextrin transport. J Bacteriol. 140:1-13.
- 49. Wang, F., X. Xiao, A. Saito, and H. Schrempf. 2002. Streptomyces olivaceoviridis possesses a phosphotransferase system that mediates specific, phosphoenolpyruvate-dependent uptake of N-acetylglucosamine. Mol Genet Genomics. 268:344-351.
- 50. Wiseman, T., S. Williston, J.F. Brandts, and L.N. Lin. 1989. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 179:131-137.
- 51. Xiao, X., F. Wang, A. Saito, J. Majaka, A. Schlösser, and H. Schrempf. 2002. The novel Streptomyces olivaceoviridis ABC transporter Ngc mediates uptake of N-acetylglucosamine and N,N′-diacetylchitobiose. Mol Genet Genomics. 267:429-439.
- 52. Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 33:103-119.






