Abstract
Microbial rhodopsins, comprising a protein moiety (rhodopsin apoprotein) bound to the light-absorbing chromophore retinal, function as ion pumps, ion channels, or light sensors. However, recent genomic and metagenomic surveys showed that some rhodopsin-possessing prokaryotes lack the known genes for retinal biosynthesis. Since rhodopsin apoproteins cannot absorb light energy, rhodopsins produced by prokaryotic strains lacking genes for retinal biosynthesis are hypothesized to be non-functional in cells. In the present study, we investigated whether Aurantimicrobium minutum KNCT, which is widely distributed in terrestrial environments and lacks any previously identified retinal biosynthesis genes, possesses functional rhodopsin. We initially measured ion transport activity in cultured cells. A light-induced pH change in a cell suspension of rhodopsin-possessing bacteria was detected in the absence of exogenous retinal. Furthermore, spectroscopic analyses of the cell lysate and HPLC-MS/MS analyses revealed that this strain contained an endogenous retinal. These results confirmed that A. minutum KNCT possesses functional rhodopsin and, hence, produces retinal via an unknown biosynthetic pathway. These results suggest that rhodopsin-possessing prokaryotes lacking known retinal biosynthesis genes also have functional rhodopsins.
Microbial rhodopsins are seven-transmembrane proteins containing a protein moiety (apoprotein) and the light-absorbing chromophore, retinal. The first member of this family of photoreceptor proteins, bacteriorhodopsin, was discovered in halophilic archaea and functions as a light-driven outward H+ pump (Oesterhelt and Stoeckenius, 1971). Microbial rhodopsin genes are widely distributed, not only among prokaryotes, but also among fungi (Brown, 2004), eukaryotic algae (Marchetti et al., 2012), and giant viruses (Yutin and Koonin, 2012). Upon illumination, the isomerization of retinal causes a conformational change in rhodopsin, and microbial rhodopsin functions as an ion pump (Oesterhelt and Stoeckenius, 1971), ion channel (Nagel et al., 2002), or light sensor (Spudich, 1998). All microbial rhodopsins are activated via light absorption by retinal, which binds to a lysine in the middle of the seventh transmembrane helix (Lys216; residue number in bacteriorhodopsin). The opsin apoprotein itself cannot receive light without a chromophore and is, thus, considered to be non-functional.
Despite the importance of retinal for functionalizing rhodopsin, the biosynthesis of this chromophore was not fully understood until 2005. A putative gene involved in the retinal biosynthetic pathway was initially discovered in the halophilic archaea, Halobacterium halobium; this gene influenced rhodopsin gene expression and was named the bacterio-opsin-related protein (brp) (Betlach et al., 1984). A bacterio-opsin-related protein homolog (blh) gene was also reported in Halobacterium sp. NRC-1 (now recognized as Halobacterium salinarum NRC-1) (Ng et al., 2000). H. salinarum NRC-1 has both brp and blh in its genome; nevertheless, the roles of these proteins remained unknown, similar to the biosynthetic pathway of retinal. In 2001, Peck et al. showed that brp and/or blh genes are required for functionalizing rhodopsin (Peck et al., 2001), although these proteins were still suggested to play a role in the transport of retinal or its binding to rhodopsin in cells, rather than in its biosynthesis. In rhodopsin-possessing prokaryotes, the retinal biosynthetic pathway was identified within the same operon as the rhodopsin gene (Sabehi et al., 2005). Sabehi et al. confirmed that the blh gene encoded a β-carotene 15,15′ dioxygenase, which cleaves β-carotene to retinal. Therefore, brp and blh were presumed to be homologs, both encoding β-carotene 15,15′ dioxygenase. Sequence homology between the brp and blh genes is so high that they cannot be clearly distinguished from each other (Nakajima et al., 2018). In contrast, in a cyanobacteria-specific pathway, retinal is produced by a 15,15′-monooxygenase encoded by the diox1 gene (Ruch et al., 2005). This monooxygenase was also shown to oxidatively cleave β-apo-carotenals in an asymmetric manner, forming retinal. Based on previous findings, rhodopsin-possessing prokaryotes are assumed to produce retinal and, thus, functional rhodopsin.
Despite these earlier theories, recent genomic and metagenomic surveys showed that some prokaryotes that possess rhodopsin genes lack a blh homolog (Dupont et al., 2012; Keffer et al., 2015; Denef et al., 2016; Pinhassi et al., 2016; Thiel et al., 2017: Table S1). Organisms such as SAR86, Marine Group II, and CL500-11 are considered to possess rhodopsin genes and are abundant in certain aquatic environments (Morris et al., 2002; Iverson et al., 2012; Okazaki et al., 2012); therefore, these rhodopsin-possessing yet blh-lacking prokaryotes appear to be common in the natural environment. A physiological study reported that the rhodopsin-possessing Actinobacteria, Rhodoluna lacicola MWH-Ta8T (Hahn et al., 2014), which lacks the blh gene, did not exhibit ion-transporting activity until its culture medium was supplied with exogenous retinal (Keffer et al., 2015). Therefore, the rhodopsin present in R. lacicola was considered to be non-functional and the strain appeared to obtain retinal from the surrounding environment. However, it currently remains unclear whether all prokaryotes that lack blh and diox1 are unable to endogenously produce retinal.
We herein conducted a functional analysis of a rhodopsin and chemical analyses of pigments from the cosmopolitan freshwater Actinobacterium isolate, Aurantimicrobium minutum KNCT (Nakai et al., 2015), which belongs to the same family as R. lacicola. The A. minutum genome was reported in 2016 (Nakai et al., 2016); it contains a gene that encodes a putative light-driven, proton-pumping rhodopsin, Xanthorhodopsin (Balashov et al., 2005) -like rhodopsin (XLR), but lacks the blh gene. We measured XLR proton-pumping activity and performed a spectroscopic analysis and high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) to clarify whether A. minutum produces retinal for the construction of a functional rhodopsin in the absence of the blh gene.
Materials and Methods
Strain and growth conditions for A. minutum KNCT
A. minutum KNCT was originally isolated from a river in Japan (Nakai et al., 2015). The strain used in the present study was provided by the Biological Resource Center, NITE (NBRC) (Chiba, Japan). A. minutum cells were incubated in 300 mL of medium (1 g L–1 Bacto Nutrient Broth, 1 g L–1 Bacto Tryptone, and 1 g L–1 Bacto Yeast extract [BD Bioscience]) modified from NSY medium (Hahn et al., 2003) under light (approximately 35 μmol photons s–1 m–2) or dark conditions at 25°C for 20 days. Cell growth was measured based on assessments of optical density at 660 nm (OD660) every 2 days using a spectrophotometer (Digital Colorimeter AC-114; Optima). After an incubation for 10 days, cells were centrifuged in preparation for the measurement of XLR proton-pumping activity and subsequent analyses.
Measurement of light-driven proton-pumping activity
Light-driven proton-pumping activity was measured according to the method described by Yoshizawa et al. (2012), with the following modifications. Cells in approximately 120–160 mL of modified NSY medium were collected by centrifugation at 7,000×g at 4°C for 8 min. Cells were then washed three times with 100 mM NaCl and resuspended in 6 mL of 100 mM NaCl. The light source was a 300-W xenon lamp (MAX-303; Asahi Spectra), and 6 mL of the cell suspension was initially placed in the dark and then irradiated for 5 min through 450-±10-nm, 520-±10-nm, and 580-±10-nm bandpass filters (MX0450, MX0520, and MX0580; Asahi Spectra). The light intensity of each wavelength was fixed at approximately 7 mW cm–2 using an optical power meter (#3664; Hioki) with an optical sensor (#9742; Hioki). pH was measured using a pH meter (LAQUA F-72; Horiba). Proton-pumping activity was defined as the slope in the 30 s following the onset of illumination. This initial slope was normalized to that obtained prior to illumination. The first measurement was performed without external retinal, and the second measurement was conducted after the addition of retinal dissolved in 100% ethanol (at a final concentration of 20 μM). All measurements were performed at 4°C.
Gene preparation and ion transport measurements of XLR in Escherichia coli cells
The A. minutum DNA fragment encoding XLR (named AmXLR; WP_096383469.1) was chemically synthesized with codon optimization for E. coli by Eurofins Genomics. This gene fragment was inserted into the NdeI and XhoI sites of the pET21a vector (Novagene); consequently, the plasmid encoded AmXLR with hexahistidine at the C terminus. The plasmid was transformed into E. coli strain C41 (DE3) (Lucigen). E. coli cells with the plasmid were initially incubated at 37°C on an LB-medium agar plate supplemented with ampicillin (final concentration, 100 μg mL–1), and were then incubated at 37°C in 100 mL of 2×YT medium supplemented with ampicillin (final concentration, 100 μg mL–1) as a pre-culture. After pre-culturing, cells were incubated at 37°C in 100 mL of modified NSY medium, which was also used for the incubation of A. minutum KNCT. Protein expression was induced by adding 0.2 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) when OD660 reached approximately 0.4. All-trans retinal (Sigma-Aldrich) was added to one flask (final concentration 10 μM; cell-retinal [+]) and not to the other flask (cell-retinal [–]). Rhodopsin-expressing cells were collected by centrifugation (4,200×g for 3 min), washed three times, and then re-suspended in 100 mM NaCl. A 300-W MAX-303 xenon lamp was used as the light source, and 6 mL of the cell suspension was initially placed in the dark and then irradiated through the 520-±10-nm bandpass filter for 5 min. pH was monitored using a LAQUA F-72 pH meter. The experiment was repeated using cells incubated with retinal under the same conditions after the addition of the protonophore CCCP (final concentration, 30 μM) to inhibit proton transport. All measurements were performed at 4°C.
Spectroscopic analysis of XLR in A. minutum cells
The A. minutum cells used for the spectroscopic analysis were incubated under the same conditions as those outlined above for the measurement of proton-pumping activity. Approximately 300 mL of modified NSY medium was concentrated by centrifugation at 7,000×g at 4°C for 8 min. Half of the resulting pellet was suspended in buffer containing 50 mM Tris-HCl and 0.1 M NaCl (pH 7.0) and then ultrasonicated for 80 cycles of 1 min on and 1 min off (UD-211; TOMY Seiko). The supernatant was then obtained by centrifugation at 35,600×g at 4°C for 10 min, followed by ultra-centrifugation at 178,000×g at 4°C for 30 min (himac CP56G; Hitachi Koki, P50A2 rotor; Hitachi Koki) to separate the pellet containing the membrane fraction. The pellet was homogenized on ice and suspended in Tris-HCl buffer at pH 7.0. Hydroxylamine was added to the suspension to a final concentration of 100 mM, and absorption spectra were measured every 10 min using the ultraviolet-visible spectrophotometer UV-2450 with the ISR2200 integrating sphere (Shimadzu). The reaction constant of the reaction, τ, was estimated from a single exponential fit:
y=y0+A*exp(–x/τ)
where y and x are the absorption change and reaction time, respectively.
HPLC-MS/MC analyses
A. minutum cells used in the HPLC-MS/MS analysis were incubated under the same conditions as those described above. Cells in approximately 300 mL of culture medium were pelleted by centrifugation at 7,000×g at 4°C for 8 min, and the supernatant was completely removed. In the HPLC-MS/MS analysis, aliquots of acetone were added to the frozen pellet in a microtube, which was then placed in an ice-cooled ultrasonication bath for pigment extraction. Samples were then ultrasonically homogenized for a few minutes, and acetone supernatants were immediately separated from the particulate material by centrifugation and directly injected into the HPLC apparatus for analysis.
The HPLC-MS/MS instrument consisted of a Shimadzu Nexera X2 liquid chromatography system, comprising a CBM-20A communication bus module, two DGU-20A3R/5R HPLC degassing units, three LC-30AD solvent delivery units constituting a ternary pumping system, an SIL-30AC autosampler, CTO-20AC column oven, and LCMS-8030 triple quadrupole mass spectrometer connected through an atmospheric pressure chemical ionization (APCI) interface (Shimadzu). The system was then coupled to a personal computer configured to run Shimadzu LabSolution software. Reverse-phase HPLC was performed under the following conditions: column, Zorbax Eclipse Plus C18 (Rapid Resolution HT, 3.0×100 mm, 1.8-μm silica particle size; Agilent Technologies); eluent, the ternary gradient program summarized in Table S2; and flow rate, 0.5 mL min–1. All mobile phases were degassed in vacuo using ultrasonication. Mobile-phase reservoir bottles were designed to prevent any contact between the mobile phases and air during the analysis. The solvents used for the HPLC mobile phases included LC/MS-grade distilled water and methanol purchased from Kanto Chemical and LC/MS-grade formic acid and HPLC-grade acetone purchased from Waco Pure Chemical Industries. APCI was set to the following conditions: nebulizer gas flow, 2.2 L min–1; interface temperature, 320°C; desolvation line temperature, 140°C; heat-block temperature, 250°C; drying gas flow, 3 L min–1. The parameters of the Q1 scan in the positive ion mode of the mass spectrometer were set as follows: scan range, m/z 100.00–400.00; event time, 0.200. Parameters for the detection of retinal during multiple reaction monitoring (MRM) in the positive ion mode were as follows: precursor ion ([M+H]+), m/z 285.25; product ion, m/z 160.10; dwell time, 10 ms; collision energy (CE), –10 V.
Results and Discussion
XLR proton-pumping activities of A. minutum with and without external retinal
A. minutum cells were incubated for 20 days under light or dark conditions. No significant differences were observed in the growth of A. minutum between the light and dark conditions until the mid-log phase (Fig. S1). Maresca et al. (2019) reported that Aurantimicrobium spp. grow more rapidly in the light than in the dark. The discrepancy between the present results from their findings may be due to differences in the medium components, the strain phenotype, or both. A. minutum cells incubated for 10 days were used to measure light-driven proton-pumping activities because the maximum OD660 during the incubation period was observed at 10 days; no significant differences were observed in OD660 between the light and dark conditions.
Light-induced pH changes in suspensions of A. minutum cells were measured using cells incubated in the light (cell-LIGHT) or dark (cell-DARK). A light-induced pH change was observed in both cell-LIGHT and cell-DARK samples in the absence of exogenous retinal (Fig. 1A) despite the absence of blh in the genome of A. minutum. To exclude the possibility that the chromophore was supplied by modified NSY medium, we measured proton-pumping activities using E. coli cells that heterologously expressed AmXLR, and a light-induced pH change was not detected without retinal (Fig. S2; gray line “Retinal [–]”). In contrast, a light-induced pH change was detected in AmXLR-expressing E. coli cells when retinal was added to the medium (Fig. S2; orange line “Retinal [+]”). These results suggest the following: (1) AmXLR expressed in E. coli functions as a light-driven proton pump if retinal is externally supplied, (2) E. coli does not produce retinal, and (3) modified NSY medium did not contain retinal. Therefore, A. minutum constructs a functional rhodopsin, the XLR apoprotein of which binds to an endogenous chromophore.

When retinal was externally supplied, a marked decrease in pH was induced by light in cell-LIGHT (Fig. 1B; orange line), whereas a markedly smaller reduction was observed in pH in cell-DARK (Fig. 1B; black line). The addition of the protonophore, CCCP, to the same cells abolished light-induced pH changes (Fig. 1C; orange and black lines). Furthermore, the light-induced pH change at green light (520 nm) was larger than those at orange (580 nm) and blue (450 nm) lights (Fig. S3). In other words, proton-pumping activity was the strongest at a wavelength close to the absorption maximum of rhodopsin. These results showed that pH changes were due to the light-driven proton pumping of XLR in A. minutum. In the analysis of XLR proton-pumping activity in cells incubated for 10 days, the initial slope of cell-LIGHT with an external supply of retinal was significantly higher than that of cell-DARK with an external supply of retinal (Fig. S4). Based on equal photon flux, proton-pumping activity was presumably proportional to the number of rhodopsin molecules activated by the binding of externally supplied retinal. Each rhodopsin molecule contains only a single retinal molecule; therefore, the quantity of retinal supplied to the cell suspension (20 mM) was excessive, and the increase observed in proton-pumping activity must have reflected the quantity of the XLR apoprotein expressed prior to the addition of retinal. Therefore, these results indicated that more XLR was expressed under light conditions than under dark conditions. Nevertheless, it is important to note that a significant difference remained between the proton-pumping activity in cell-LIGHT with and without externally supplied retinal (Fig. S4). These results suggested that some, but not all, XLR apoproteins expressed in cell-LIGHT were functionalized by accepting some chromophores that must have been endogenously supplied by A. minutum. In other words, A. minutum may produce a smaller amount of the chromophore than the XLR apoprotein, such that the amount of functional XLR in A. minutum is limited by the concentration of the endogenous chromophore, possibly retinal, and some of the XLR expressed is non-functional. In the rhodopsin-possessing bacterium, Vibrio sp. AND4, light-enhanced long-term survival during starvation was suggested to have been mediated by proton-pumping rhodopsin (Gómez-Consarnau et al., 2010; Akram et al., 2013). Proton-pumping rhodopsin may generate ATP under irradiation with light and reduce the consumption of organic matter and/or oxygen in the medium used for energy metabolism. Once the bacterium maintains the same metabolic activity, the conservation of organic matter via this rhodopsin function is considered to prolong the survival of the bacterium (Steindler et al., 2011). The production of the chromophore by A. minutum may be up-regulated under these conditions, such that the utilization of rhodopsin becomes more important for its survival. Therefore, similar to other blh-lacking prokaryotes, native A. minutum may utilize retinal externally supplied by other retinal producers inhabiting the same environment if it becomes available.
Spectroscopic analysis of native A. minutum cells after hydroxylamine bleaching
We performed a spectroscopic analysis to examine whether A. minutum produces retinal as an endogenous chromophore of XLR. The spectra from cell-LIGHT and cell-DARK were obtained from the lysate after the addition of hydroxylamine. In the presence of retinal-bound rhodopsin, an absorbance peak of retinal oxime was expected to emerge upon the bleaching of rhodopsin (Sudo et al., 2002). As shown in Fig. 2, the absorbance intensity at the AmXLR absorption maximum (approximately 532 nm) decreased, whereas that at the retinal oxime maximum (approximately 363 nm) increased. Furthermore, τ of cell-LIGHT (τ-light) was estimated at 481-nm datasets, while τ of cell-DARK (τ-dark) was estimated at 483-nm datasets because these wavelengths are the peak wavelengths of the different absorptions (Fig. 2C). τ-light and τ-dark were approximately 1,300 min (22 h) and 1,400 min (23 h), respectively. Since the time constant of the reaction of BR was estimated to be approximately 10 h (Subramaniam et al., 1991), τ-light and τ-dark were not markedly different from τ of BR.
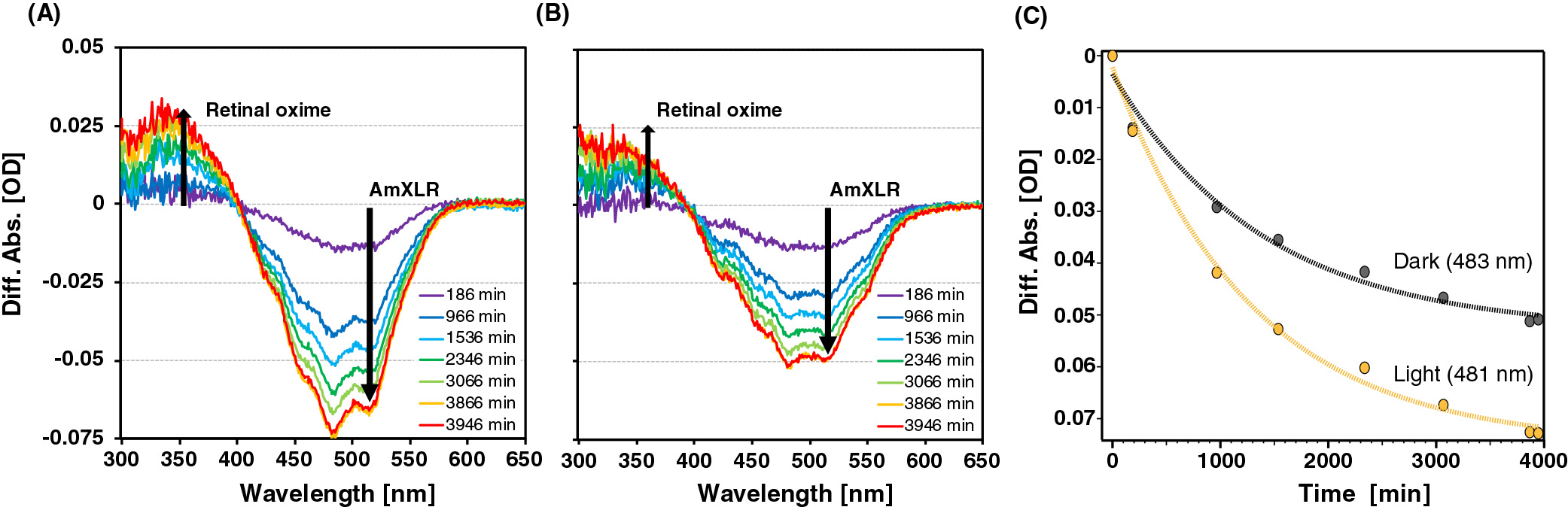
These results suggest that rhodopsin underwent discoloration by desorbing retinal oxime, indicating that at least some XLR in A. minutum KNCT bound retinal. However, these differences in spectra may have resulted from a chromophore with a formyl group that is analogous to retinal. Therefore, we performed HPLC-MS/MS analyses to establish whether the chromophore produced by A. minutum is retinal.
Identification of endogenous retinal by the HPLC-MS/MS analysis
We obtained the product ion spectrum of the authentic standard of all-trans retinal on the expected precursor mass [M+H]+ with m/z=285.3, identifying the primary ion product with m/z=161.2, which was consistent with the theoretical value m/z=161.1 (Fig. 3A). Unique peaks with identical retention times were observed in the mass spectra on the expected MRM transition of retinal [m/z=285.3>m/z=161.1] of both the authentic standard and A. minutum extract (Fig. 3B), suggesting the occurrence of retinal in A. minutum KNCT. Furthermore, the fragmentation pattern of the product ion spectrum on the expected precursor mass was obtained at the identified peak of the A. minutum extract (Fig. 3C) and matched well with the pattern of the authentic standard (Fig. 3A), confirming the detection of retinal in the bacterial extract. On the other hand, we did not detect any carotenoid signals, which potentially are retinal precursors, under the HPLC-MS/MS conditions used in the present study.

Microbial rhodopsins have several retinal isomers, such as all-trans, 13-cis, 11-cis, and 9-cis. Of these, most microbial rhodopsins (particularly proton-pumping rhodopsins) only work with all-trans retinal. Therefore, we assumed that AmXLR has an all-trans retinal as a chromophore. However, Fig. S4 shows that this strain did not produce sufficient amounts of retinal capable of binding to all XLR apoproteins. Since only two culture conditions (light and dark) were employed in the present study, it currently remains unclear whether A. minutum produced sufficient retinal under different culture conditions. Peck et al. (2001) showed that the amount of functional rhodopsin (bacteriorhodopsin) increased following the addition of exogenous retinal in H. salinarum. This increase was greater in a brp gene deletant (that only possessed the blh gene), indicating that not all rhodopsin are functional, even though this archaeon produces retinal. In other words, the amount of retinal may be less than that of rhodopsin apoproteins among some rhodopsin-possessing prokaryotes.
Hypothesis of an unknown retinal biosynthetic pathway
Although we showed that A. minutum KNCT produced retinal, the genes responsible for its biosynthesis have not yet been identified. Among rhodopsin-possessing Actinobacteria, a lycopene cyclase encoded by the crtYc/Yd genes catalyzes the conversion of lycopene to β-carotene, which is then supplied for endogenous retinal (Dwulit-Smith et al., 2018). However, A. minutum has putative crtYe/Yf cluster genes, which encode C50 carotenoid epsilon cyclase (Krubasik et al., 2001; Netzer et al., 2010), not crtYc/Yd genes. Representative crtYe/Yf genes are found in Corynebacterium glutamicum (accession numbers: AUI03847 and AUI03848) and Micrococcus luteus (accession numbers: AYO50480 and AYO50481), to which the putative genes of A. minutum are closely related; however, they are less closely related to the crtYc/Yd clusters (Fig. S5). It has not yet been established whether C50 carotenoid epsilon cyclase also converts lycopene (C40 carotenoid) to β-carotene; therefore, A. minutum KNCT, similar to other rhodopsin-possessing Actinobacteria, may not produce β-carotene from lycopene. In contrast, A. minutum has a putative crtEb gene encoding lycopene elongase, which converts lycopene to a C50 carotenoid. C50 carotenoid epsilon cyclase encoded by the crtYe/Yf genes may form cyclic ends to convert a β-carotene analog of the C50 carotenoid with an extended isoprenyl chain. Since A. minutum possesses genes involved in C50 carotenoid biosynthesis, we hypothesized that this strain may have an oxygenase that asymmetrically cleaves cyclic C50 carotenoid to produce all-trans retinal (Fig. S6). Furthermore, according to the phylogenetic tree shown in Fig. S5, R. lacicola MWH-Ta8T, which was not considered to produce retinal, at least possesses the crtYe/Yf genes (and also a crtEb gene). Therefore, R. lacicola may also produce retinal through an unknown pathway similar to that in A. minutum. However, the present results do not exclude the possibility of a retinal biosynthesis pathway without the C50 carotenoid as an intermediate. While a β-carotene 15,15′ dioxygenase cleaves one β-carotene to two retinal, asymmetrical cleavage may convert one C50 carotenoid to one retinal, such as 15,15′-monooxygenase, encoded by the diox1 gene. In that case, the number of retinal molecules produced by asymmetrical cleavage is half the number of retinal molecules produced by dioxygenase cleavage. This may be one of the reasons why A. minutum only functionalizes some of the XLR expressed.
In conclusion, we herein report that some XLR rhodopsins in the freshwater Actinobacterium, A. minutum KNCT, which possesses a rhodopsin gene and lacks the known retinal biosynthetic genes blh and diox1, are functional. The cleavage of β-carotene is not an abiotic reaction, but an enzymatic reaction that produces retinal (Olson and Hayaishi, 1965); therefore, A. minutum KNCT must produce (all-trans) retinal using an unknown retinal biosynthetic gene(s) for functionalizing rhodopsin. Although rhodopsins in prokaryotes that lack blh or diox1 were previously considered to be non-functional, the endogenous production of retinal was confirmed in this strain. Interestingly, diverse organisms that include strains belonging to the phyla Deinococcus-Thermus, Firmicutes, Proteobacteria, Bacteroidetes, Chloroflexi, and Euryarchaeota have putative rhodopsin genes, but lack blh and diox1 (data not shown); however, it currently remains unclear whether these organisms endogenously produce retinal without the currently known retinal biosynthetic genes. Further studies on these strains may reveal how widespread retinal production is in blh-lacking strains and also how these rhodopsin-possessing prokaryotes obtain retinal to construct functional rhodopsins.
Citation
Nakajima, Y., Kojima, K., Kashiyama, Y., Doi, S., Nakai, R., Sudo, Y., et al. (2020) Bacterium Lacking a Known Gene for Retinal Biosynthesis Constructs Functional Rhodopsins. Microbes Environ 35: ME20085.
https://doi.org/10.1264/jsme2.ME20085
Acknowledgements
Aurantimicrobium minutum KNCT (=NBRC 105389T) was obtained from the Biological Resource Center, NITE (NBRC). This work was supported by the Japan Society for the Promotion of Science (JSPS) to YN (JP19J01352), YK (JP17H03723, JP18H03743), YS (JP18H02411, JP19H05396 and JP19H04727), RN (JP15H05620), and SY (JP18H04136).
References
- Akram, N., Palovaara, J., Forsberg, J., Lindh, M.V., Milton, D.L., Luo, H., et al. (2013) Regulation of proteorhodopsin gene expression by nutrient limitation in the marine bacterium Vibrio sp. AND 4. Environ Microbiol
15: 1400–1415.
- Balashov, S.P., Imasheva, E.S., Boichenko, V.A., Antón, J., Wang, J.M., and Lanyi, J.K. (2005) Xanthorhodopsin: a proton pump with a light-harvesting carotenoid antenna. Science
309: 2061–2064.
- Betlach, M., Friedman, J., Boyer, H.W., and Pfeifer, F. (1984) Characterization of a halobacterial gene affecting bacterio-opsin gene expression. Nucleic Acids Res
12: 7949–7959.
- Brown, L.S. (2004) Fungal rhodopsins and opsin-related proteins: eukaryotic homologues of bacteriorhodopsin with unknown functions. Photochem Photobiol Sci
3: 555–565.
- Denef, V.J., Mueller, R.S., Chiang, E., Liebig, J.R., and Vanderploeg, H.A. (2016) Chloroflexi CL500-11 populations that predominate deep-lake hypolimnion bacterioplankton rely on nitrogen-rich dissolved organic matter metabolism and C1 compound oxidation. Appl Environ Microbiol
82: 1423–1432.
- Dupont, C.L., Rusch, D.B., Yooseph, S., Lombardo, M.J., Richter, R.A., Valas, R., et al. (2012) Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J
6: 1186–1199.
- Dwulit-Smith, J.R., Hamilton, J.J., Stevenson, D.M., He, S., Oyserman, B.O., Moya-Flores, F., et al. (2018) acI Actinobacteria assemble a functional actinorhodopsin with natively synthesized retinal. Appl Environ Microbiol
84: e01678-18.
- Gómez-Consarnau, L., Akram, N., Lindell, K., Pedersen, A., Neutze, R., Milton, D.L., et al. (2010) Proteorhodopsin phototrophy promotes survival of marine bacteria during starvation. PLoS Biol
8: e1000358.
- Hahn, M.W., Lünsdorf, H., Wu, Q., Schauer, M., Höfle, M.G., Boenigk, J., and Stadler, P. (2003) Isolation of novel ultramicrobacteria classified as Actinobacteria from five freshwater habitats in Europe and Asia. Appl Environ Microbiol
69: 1442–1451.
- Hahn, M.W., Schmidt, J., Taipale, S.J., Doolittle, W.F., and Koll, U. (2014) Rhodoluna lacicola gen. nov., sp. nov., a planktonic freshwater bacterium with stream-lined genome. Int J Syst Evol Microbiol
64: 3254.
- Iverson, V., Morris, R.M., Frazar, C.D., Berthiaume, C.T., Morales, R.L., and Armbrust, E.V. (2012) Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science
335: 587–590.
- Keffer, J.L., Hahn, M.W., and Maresca, J.A. (2015) Characterization of an unconventional rhodopsin from the freshwater Actinobacterium Rhodoluna lacicola. J Bacteriol
197: 2704–2712.
- Krubasik, P., Kobayashi, M., and Sandmann, G. (2001) Expression and functional analysis of a gene cluster involved in the synthesis of decaprenoxanthin reveals the mechanisms for C50 carotenoid formation. Eur J Biochem
268: 3702–3708.
- Marchetti, A., Schruth, D.M., Durkin, C.A., Parker, M.S., Kodner, R.B., Berthiaume, C.T., et al. (2012) Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability. Proc Natl Acad Sci U S A
109: E317–E325.
- Maresca, J.A., Keffer, J.L., Hempel, P., Polson, S.W., Shevchenko, O., Bhavsar, J., et al. (2019) Light modulates the physiology of non-phototrophic Actinobacteria. J Bacteriol
201: e00740-18.
- Morris, R.M., Rappé, M.S., Connon, S.A., Vergin, K.L., Siebold, W.A., Carlson, C.A., and Giovannoni, S.J. (2002) SAR11 clade dominates ocean surface bacterioplankton communities. Nature
420: 806–810.
- Nagel, G., Ollig, D., Fuhrmann, M., Kateriya, S., Musti, A.M., Bamberg, E., and Hegemann, P. (2002) Channelrhodopsin-1: a light-gated proton channel in green algae. Science
296: 2395–2398.
- Nakai, R., Baba, T., Niki, H., Nishijima, M., and Naganuma, T. (2015) Aurantimicrobium minutum gen. nov., sp. nov., a novel ultramicrobacterium of the family Microbacteriaceae, isolated from river water. Int J Syst Evol Microbiol
65: 4072–4079.
- Nakai, R., Fujisawa, T., Nakamura, Y., Nishide, H., Uchiyama, I., Baba, T., et al. (2016) Complete genome sequence of Aurantimicrobium minutum type strain KNCT, a planktonic ultramicrobacterium isolated from river water. Genome Announc
4: e00616-16.
- Nakajima, Y., Tsukamoto, T., Kumagai, Y., Ogura, Y., Hayashi, T., Song, J., et al. (2018) Presence of a haloarchaeal halorhodopsin-like Cl– pump in marine bacteria. Microbes Environ
33: 89–97.
- Netzer, R., Stafsnes, M.H., Andreassen, T., Goksøyr, A., Bruheim, P., and Brautaset, T. (2010) Biosynthetic pathway for γ-cyclic sarcinaxanthin in Micrococcus luteus: Heterologous expression and evidence for diverse and multiple catalytic functions of C50 carotenoid cyclases. J Bacteriol
192: 5688–5699.
- Ng, W.V., Kennedy, S.P., Mahairas, G.G., Berquist, B., Pan, M., Shukla, H.D., et al. (2000) Genome sequence of Halobacterium species NRC-1. Proc Natl Acad Sci U S A
97: 12176–12181.
- Oesterhelt, D., and Stoeckenius, W. (1971) Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nature
233: 149–152.
- Okazaki, Y., Hodoki, Y., and Nakano, S.I. (2012) Seasonal dominance of CL500-11 bacterioplankton (phylum Chloroflexi) in the oxygenated hypolimnion of Lake Biwa, Japan. FEMS Microbiol Ecol
83: 82–92.
- Olson, J.A., and Hayaishi, O. (1965) The enzymatic cleavage of beta-carotene into vitamin A by soluble enzymes of rat liver and intestine. Proc Natl Acad Sci U S A
54: 1364.
- Peck, R.F., Echavarri-Erasun, C., Johnson, E.A., Ng, W.V., Kennedy, S.P., Hood, L., et al. (2001) brp and blh are required for synthesis of the retinal cofactor of bacteriorhodopsin in Halobacterium salinarum. J Biol Chem
276: 5739–5744.
- Pinhassi, J., DeLong, E.F., Béjà, O., González, J.M., and Pedrós-Alió, C. (2016) Marine bacterial and archaeal ion-pumping rhodopsins: genetic diversity, physiology, and ecology. Microbiol Mol Biol Rev
80: 929–954.
- Ruch, S., Beyer, P., Ernst, H., and Al-Babili, S. (2005) Retinal biosynthesis in Eubacteria: in vitro characterization of a novel carotenoid oxygenase from Synechocystis sp. PCC 6803. Mol Microbiol
55: 1015–1024.
- Sabehi, G., Loy, A., Jung, K.H., Partha, R., Spudich, J.L., Isaacson, T., et al. (2005) New insights into metabolic properties of marine bacteria encoding proteorhodopsins. PLoS Biol
3: e273.
- Spudich, J.L. (1998) Variations on a molecular switch: transport and sensory signalling by archaeal rhodopsins. Mol Microbiol
28: 1051–1058.
- Steindler, L., Schwalbach, M.S., Smith, D.P., Chan, F., and Giovannoni, S.J. (2011) Energy starved Candidatus Pelagibacter ubique substitutes light-mediated ATP production for endogenous carbon respiration. PLoS One
6: e19725.
- Subramaniam, S., Marti, T., Rösselet, S.J., Rothschild, K.J., and Khorana, H.G. (1991) The reaction of hydroxylamine with bacteriorhodopsin studied with mutants that have altered photocycles: selective reactivity of different photointermediates. Proc Natl Acad Sci U S A
88: 2583–2587.
- Sudo, Y., Iwamoto, M., Shimono, K., and Kamo, N. (2002) Association of pharaonis phoborhodopsin with its cognate transducer decreases the photo-dependent reactivity by water-soluble reagents of azide and hydroxylamine. Biochim Biophys Acta
1558: 63–69.
- Thiel, V., Hügler, M., Ward, D.M., and Bryant, D.A. (2017) The dark side of the Mushroom Spring microbial mat: life in the shadow of chlorophototrophs. II. Metabolic functions of abundant community members predicted from metagenomic analyses. Front Microbiol
8: 943.
- Yoshizawa, S., Kawanabe, A., Ito, H., Kandori, H., and Kogure, K. (2012) Diversity and functional analysis of proteorhodopsin in marine Flavobacteria. Environ Microbiol
14: 1240–1248.
- Yutin, N., and Koonin, E.V. (2012) Proteorhodopsin genes in giant viruses. Biol Direct
7: 34.



