Abstract
Isolated RNA viruses mainly parasitize eukaryotes. RNA viruses either expand horizontally by infecting hosts (acute type) or coexist with the host and are vertically inherited (persistent type). The significance of persistent-type RNA viruses in environmental viromes (the main hosts are expected to be microbes) was only recently reported because they had previously been overlooked in virology. In this review, we summarize the host-virus relationships of eukaryotic microbial RNA viruses. Picornavirales and Reoviridae are recognized as representative acute-type virus families, and most of the microbial viruses in Narnaviridae, Totiviridae, and Partitiviridae are categorized as representative persistent-type viruses. Acute-type viruses have only been found in aquatic environments, while persistent-type viruses are present in various environments, including aquatic environments. Moreover, persistent-type viruses are potentially widely spread in the RNA viral sequence space. This emerging evidence provides novel insights into RNA viral diversity, host-virus relationships, and their history of co-evolution.
An RNA virus has single- or double-stranded RNA as its genome. The genome sizes of RNA viruses range from several kb to several tens of kb, and some harbor segmented genomes depending on the virus group/species (King et al., 2012; Koonin et al., 2020). To date, most RNA viruses have been isolated from eukaryotes, and more than 50% of isolated viruses from eukaryotes are RNA viruses (Nasir et al., 2014). Since the majority of eukaryotes are expected to be infected by RNA viruses, RNA viruses and eukaryotes likely have a long history of co-evolution (Koonin et al., 2015). In contrast, no or very few RNA virus families have been isolated from archaea and bacteria, respectively; however, the diversity of prokaryotic RNA viruses was very recently expanded by metatranscriptomics and subsequent in silico analyses (Callanan et al., 2020; Neri, U., et al. 2022. A five-fold expansion of the global RNA virome reveals multiple new clades of RNA bacteriophages. bioRxiv
https://doi.org/10.1101/2022.02.15.480533).
Historically, RNA viruses have been detected as infectious causative agents in humans and economically important plants and animals (Beijerinck, 1898; Loeffler and Frosch, 1898; Reed and Carroll, 1901). However, in the last several decades, fungal RNA viruses that persistently infect their hosts without causing clear phenotypic changes have been discovered, although a few have been found in edible mushrooms and pathogenic fungi that cause phenotypic symptoms (Nuss, 2005; Roossinck, 2014). Moreover, similar “cryptic” RNA viruses are widely present in plants (Roossinck et al., 2010). To obtain a more detailed understanding of RNA viruses, Roossinck categorized the lifestyles of plant/fungal viruses into two types, persistent and acute (Roossinck, 2010, 2012, 2015). Briefly, viruses with a persistent lifestyle are transmitted vertically via cell division, while viruses with an acute lifestyle are most frequently transmitted horizontally (Fig. 1). This classification is completely different from “acute infection”, “persistent (latent) infection”, and “chronic infection” in human infections. These clinical categories indicate the course of viral dose/activity or the state of disease in humans. For example, hepatitis C virus (HCV), a positive-sense single-stranded RNA virus, sometimes establishes a chronic infection after an acute infection. However, this does not represent a change in the viral lifestyle. HCV is categorized as an acute-type RNA virus by Roossinck’s criteria because it mainly spreads by horizontal transmission.

The acute and persistent types have both been identified in eukaryotic microbial lineages of fungi and protists. Furthermore, viral metagenomics enables us to identify diverse RNA viruses and infer their lifestyles. To date, a large diversity of RNA viruses has been revealed in soil and aquatic microbial ecosystems (Culley et al., 2006; Suttle, 2016; Urayama et al., 2018a; Starr et al., 2019; Wolf et al., 2020; Edgar et al., 2022; Zayed et al., 2022), in addition to biological samples from organisms, such as invertebrates, deep-sea animals, and microbial consortia associated with sponges, macroalgae, and lichens (Shi et al., 2016; Urayama et al., 2018b, 2020a, 2020b; Chiba et al., 2020). In this review, we summarize eukaryotic microbial RNA viruses based on their lifestyles and discuss their significance in microbial ecosystems. This review focuses on RNA viruses excluding retroviruses. Retroviruses are distinct from other RNA viruses in their life cycle because they are embedded into the host genome, whereas other RNA viruses are not. Retroviruses have a mixed strategy of vertical and horizontal transmission and change their strategy depending on the life stage.
Microbial RNA viruses have two life cycle strategies
We define acute- and persistent-type life cycles in microbial RNA viruses based on Roossinck’s definition (Roossinck, 2010) (Fig. 1). Acute-type viruses are mainly transmitted horizontally via extracellular viral particles. These viruses have the ability to enter host cells from the outside and exit host cells to the outside. During the viral life cycle, acute-type viruses often cause “disease” as represented by cell lysis (Tomaru et al., 2004). In contrast, persistent-type viruses are mainly transmitted vertically via the cell division of their host without cell lysis (Nuss, 2005). These viruses remain associated with their hosts for many generations with nearly 100% vertical transmission, and sometimes lack the ability to enter and exit host cells. In addition, some persistent-type RNA viruses lack capsid proteins (Dolja and Koonin, 2012). No gene is shared only in persistent-type RNA viruses. Persistence and vertical transmission in viruses has been suggested to correlate with commensal or mutualistic lifestyles (Roossinck, 2011; Marquez and Roossinck, 2012).
The host cell phenotype is an attractive subject in virology, and persistent- and acute-type lifestyles overlap with symbiosis and antagonism, respectively. However, difficulties are associated with the classification of viruses according to the phenotype of the host organism because the host-virus relationship changes depending on the surrounding conditions (Xu et al., 2008). Furthermore, we cannot test all environmental conditions that may affect the host phenotype. Therefore, we defined the two types of viruses based on the viral transmission route in this review.
Heterosigma akashiwo RNA virus (HaRNAV) (Tai et al., 2003; Lang et al., 2004) is a well-known acute-type RNA virus associated with microbial eukaryotes in aquatic ecosystems. HaRNAV was isolated from a Heterosigma akashiwo culture that showed the lysis of host cells after the inoculation of a virus particle fraction from seawater. To maintain HaRNAV in lab conditions, a 0.22-μm filtered cell lysate including HaRNAV is inoculated into H. akashiwo cultures, and after cell lysis, the lysate is used as an inoculum. In this case, since the host is re-infected with the daughter virus obtained by filter filtration from the lytic solution, we defined the RNA virus as an acute-type virus. Based on the genome sequence of HaRNAV, it belongs to Marnaviridae (Lang et al., 2021), which includes several acute-type microbial RNA viral genera, such as Marnavirus and Labyrnavirus (Lang et al., 2004; Takao et al., 2006).
Among persistent-type RNA viruses, Saccharomyces cerevisiae virus L-A (ScV-L-A) (Wickner, 1996) is a representative strain. The presence of RNA viruses may be confirmed by long cellular dsRNA because it only consists of dsRNA viral genomes and/or replicative intermediates of non-retro ssRNA viruses (Morris and Dodds, 1979). ScV-L-A does not confer a detectable phenotype upon its host (yeast) cells (Schmitt and Breinig, 2002), and viral dsRNA is maintained in the host cell and transmitted to daughter cells via cell division. ScV-L-A may be maintained and amplified in S. cerevisiae cells under laboratory conditions. In this case, since the RNA virus is maintained by a host sub-culture, we define the RNA virus as a persistent-type virus. ScV-L-A belongs to the family Totiviridae, which includes other known totiviruses isolated from protists (Goodman et al., 2011).
The definitions of acute- and persistent-type viruses are not applicable in some exceptional cases. For example, fungal persistent-type RNA viruses are mainly transmitted to daughter cells, but may also be transmitted between closely related fungal strains through anastomosis (Liu et al., 2003; Vainio et al., 2011). On a geological time scale, the transmission of RNA viruses between plants and fungi may also have occurred (Nibert et al., 2014; Roossinck, 2019; Bian et al., 2020).
Isolated microbial RNA viruses
We herein summarized and grouped isolated microbial RNA viruses based on the infection type (Table S1). Microbial RNA viruses with acute-type lifestyles have been obtained from aquatic host strains (Sadeghi et al., 2021). Among them, ssRNA viruses that lyse host cells have been reported, such as Heterosigma akashiwo RNA virus (HaRNAV) (Picornavirales), which infects the toxic bloom-forming raphidophyte Heterosigma akashiwo (Raphidophyceae) (Tai et al., 2003); Heterocapsa circularisquama RNA virus 01 (Alvernaviridae), which infects the bivalve-killing dinoflagellate Heterocapsa circularisquama (Dinophyceae) (Tomaru et al., 2004); and Aurantiochytrium single-stranded RNA virus 01 (Picornavirales), which infects the marine fungoide Schizochytrium sp. (Thraustochytriaceae) (Takao et al., 2005). The dsRNA virus, Micromonas pusilla reovirus (Reoviridae), which infects and lyses the marine photosynthetic protist Micromonas pusilla (Mamiellophyceae) (Brussaard et al., 2004), has also been isolated.
In contrast, microbial RNA viruses with persistent-type lifestyles have mainly been identified using intracellular dsRNA (a molecular biomarker) or RNA-seq technology. This type of RNA virus has been exclusively studied and identified in fungi (Ghabrial et al., 2015; Kotta-Loizou and Coutts, 2017). To date, identified dsRNA viruses include unclassified RNA viruses that infect the cultivated mushroom Agaricus bisporus (van der Lende et al., 1994); Penicillium chrysogenum virus (Chrysoviridae), which infects the penicillin-producing strains of Penicillium chrysogenum (Banks et al., 1969); and Saccharomyces cerevisiae virus L-A (Totiviridae), which infects the budding yeast S. cerevisiae (Wickner, 1996). Oomycetes (Oomycota) have also been subject to dsRNA-based surveillance, and the plant pathogens Phytophthora infestans and Asparagus officinalis are known to harbor Phytophthora infestans RNA virus 3 (unclassified) and Phytophthora endornavirus 2 (and 3) (Endornaviridae), respectively (Cai et al., 2013; Uchida et al., 2021). In addition, protozoans, such as Trichomonas, Leishmania, and Giardia, harbor persistent-type RNA viruses (Wang and Wang, 1986; Stuart et al., 1992; Khoshnan and Alderete, 1994); however, the presence of extracellular infection routes has been suggested (Wang and Wang, 1986; Torrecilhas et al., 2020).
RNA viruses with acute-type lifestyles have been detected in aquatic environments, while those with persistent-type lifestyles have been found in terrestrial environments. However, ssRNA and dsRNA viruses with persistent-type lifestyles have both been recently identified from a marine oomycete strain in the genus Halophytophthora (Botella et al., 2020) and a marine fungal strain isolated from the seagrass Posidonia oceanica (Nerva et al., 2016). The identification of these strains was based on high-throughput sequencing methods, while culture-dependent isolation has typically been used to identify viruses in aquatic research. However, in terrestrial environments, few attempts have been made to obtain acute-type RNA viruses. We cannot exclude the possibility that the different distribution pattern observed between the two lifestyle types is a result of methodological bias in virus detection and isolation; however, acute-type RNA viruses may have advantages in dispersal via viral particles to access new host cells in aquatic environments. In the case of DNA phages, a number of theories have been proposed, including Kill-the-Winner and Piggyback-the-Winner (Obeng et al., 2016; Pratama and van Elsas, 2018). Theoretical modeling in addition to further studies will provide more detailed insights into the distribution of acute- and persistent-type RNA viruses under diverse environmental conditions.
Persistent and acute RNA viruses in the RNA viral sequence space
To visualize the richness of persistent-type RNA viruses in the sequence space, RNA viruses isolated from eukaryotic microorganisms were classified into acute- or persistent-types based on culture-dependent laboratory observations (Table S1), as described above. In total, 96% (304/314) of eukaryotic microbial RNA viruses were recognized as persistent types (Fig. 2), while we were unable to exclude the possibility of experimental biases in culture-dependent analyses to obtain acute-type RNA viruses in eukaryotic microorganisms. To predict the distribution of persistent-type RNA viruses in the total RNA virome sequence space, we used this list as an operational reference virus list because of the lack of knowledge on persistent-type RNA viruses in other host organisms and limited metagenomic data. Based on the operational reference virus list, RNA viral clusters including all known viral RdRp sequences were analyzed, and clusters including RNA viruses related to microbial persistent-type viruses (>50% amino acid similarity) were distinguished from those of acute-type viruses (Fig. 3). In our current analysis, more than 1/3 of the clusters (including five or more sequences) were predicted to include persistent-type viruses. Since the number of isolated persistent-type RNA viruses is limited, the true richness of persistent-type RNA viruses needs to be higher than that estimated.
Microbial RNA viruses in metagenomic data
Metagenomics is a powerful tool for identifying DNA viromes in nature (Breitbart et al., 2002; Edwards and Rohwer, 2005; Suttle, 2016), and its use for RNA viromes has markedly increased (Culley et al., 2006, 2010; Steward et al., 2013; Wolf et al., 2020; Edgar et al., 2022; Zayed et al., 2022). RNA virome research is conducted as either metagenomics (metatranscriptomics) of RNA virus particles or as cellular transcriptomics. RNA virus particles in extracellular environments are expected to predominantly be acute-type viruses, and cellular RNA may harbor viral RNA from both acute- and persistent-type viruses (Fig. 1). Accordingly, cellular RNA-specific viruses are expected to be of the persistent type. In addition, dsRNA-seq for cellular RNA has also been used to detect cellular viral RNA sequences in some studies in order to more efficiently retrieve RNA viral sequences (Decker and Parker, 2014; Decker et al., 2019; Izumi et al., 2021).
In aquatic environments, most of the acute-type RNA viruses identified from extracellular viral particles belong to specific RNA viral lineages, such as Picornavirales (ssRNA) and Reoviridae (dsRNA) (Culley et al., 2006, 2014; Djikeng et al., 2009; Steward et al., 2013; Culley, 2017; Urayama et al., 2018a; Wolf et al., 2020). In addition, Picobirnaviridae (dsRNA) viruses that may infect bacterial hosts were identified as a dominant RNA virus population (Ghosh and Malik, 2021; Neri, U., et al. 2022. bioRxiv https://doi.org/10.1101/2022.02.15.480533.). The contribution of Picornavirales to viral lysis via their acute lifestyle and subsequent release of organic matter (previously defined as the virus shunt in DNA virus studies [Wilhelm and Suttle, 1999; Zimmerman et al., 2020]) was suggested based on the correlation between the relative abundance of transcripts related to Picornavirales and the amount of particulate organic carbon in pelagic ecosystems (Kaneko et al., 2021). These findings are also consistent with the results of culture-dependent isolation experiments as described above.
Based on pelagic microbial cellular RNA, in addition to Picornavirales, Reoviridae, and Picobirnaviridae, a wide range of RNA virus groups has been identified (Urayama et al., 2016, 2018a). Among them, the dominant members belong to Narnaviridae (ssRNA) and Partitiviridae (dsRNA), which have been recognized as persistent-type RNA viruses associated with fungi and plants/eukaryotic microorganisms, respectively (Hillman and Cai, 2013; Nibert et al., 2014). Moreover, RNA virome studies have been conducted on macrocolonies of eukaryotic microorganisms, such as Delisea pulchra (red algae) and Scytosiphon lomentaria (brown algae). An association with Totiviridae (dsRNA) viral genomes, which are a persistent type, is generally observed; however, diverse RNA virus groups constitute the viromes (Lachnit et al., 2016; Urayama et al., 2016; Chiba et al., 2020). In contrast, Leviviridae (ssRNA bacteriophage) and Narnaviridae virus operational taxonomic units (OTUs) have been identified as the predominant populations in soil RNA viromes, although the physical separation of cellular and viral particles has not been examined (Starr et al., 2019).
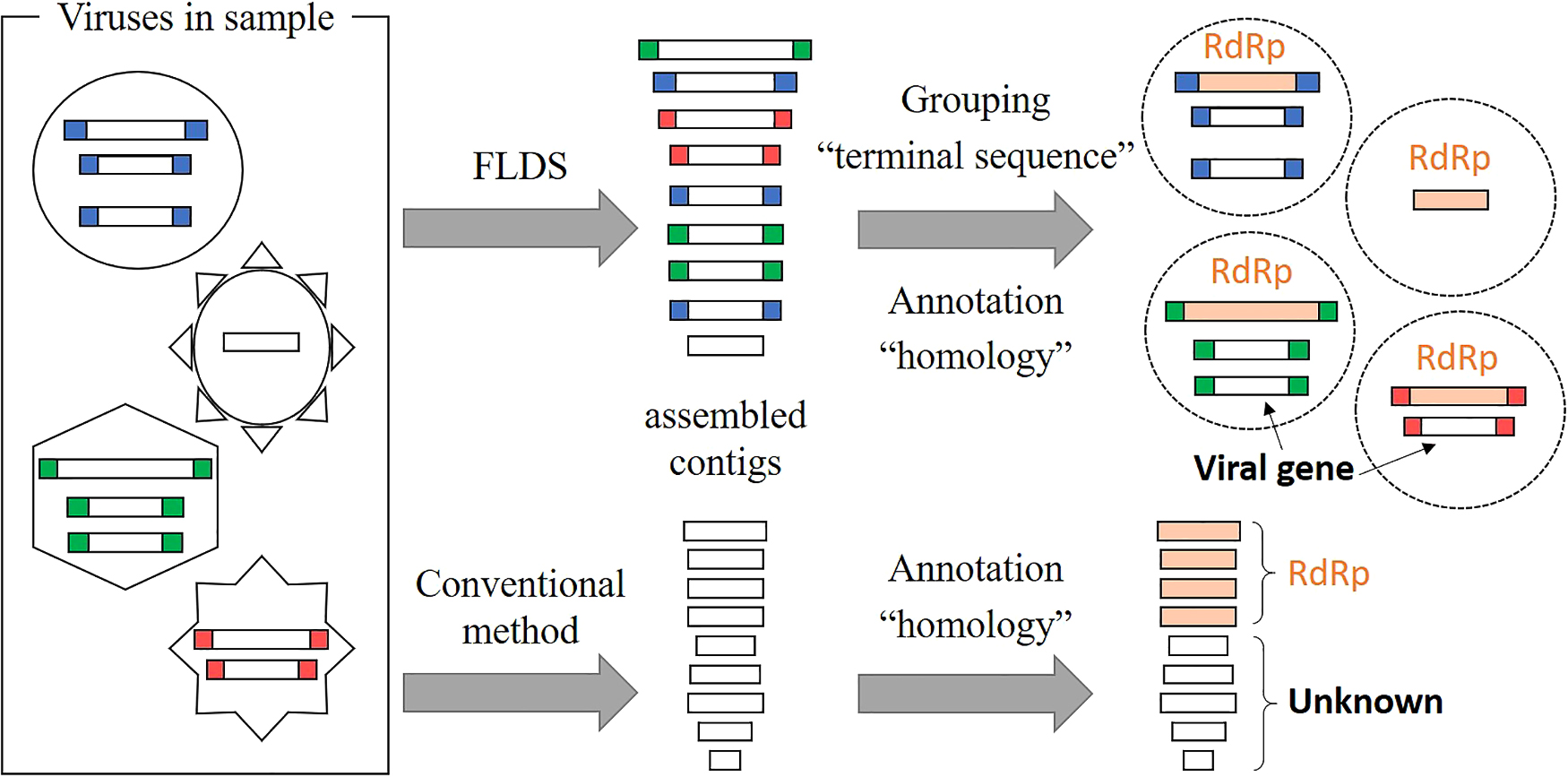
We previously compared viral dsRNA in extracellular virus particle fractions and cellular dsRNA from surface seawater (Urayama et al., 2018a). We showed 3.7- to 14.9-fold more dsRNA viral OTUs in cellular dsRNA metatranscriptomes than in extracellular dsRNA metatranscriptomes. Although we cannot rule out the possibility that some dsRNA virus particles were lost during sample processing, cellular dsRNA viruses, presumed to be of the persistent type, were abundant in the aquatic environment. In the present and related studies, we used a combination of fragmented and primer-ligated dsRNA sequencing (FLDS) and genome reconstruction in silico to identify viral sequences that are distinct from those in public databases. FLDS is a sequencing method that is applicable to long dsRNAs and enables the retrieval of the complete genomic sequences of both dsRNA and ssRNA viruses (Urayama et al., 2016; Fukasawa et al., 2020; Hirai et al., 2021; Uehara-Ichiki et al., 2021). In this technique, the terminal sequences of an RNA virus genome or genome segment may both be identified by sequence read mapping (Fig. 4). Therefore, it may be used to reconstruct multi-segmented RNA viral genomes based on terminally conserved sequences among segments (Fig. 5). In other words, a group of contigs sharing either/both terminal end sequences may represent a multi-segmented RNA virus genome. Moreover, if an RdRp gene or other virus proteins in any one of the segments in a reconstructed genome is identified based on sequence-dependent methods, all of the segments in the reconstructed genome may subsequently be identified as viral genome segments. We previously identified approximately 800 novel RNA viral genes that did not show significant (e-value less than 1×10–5) similarities to known protein sequences (Urayama et al., 2018a). Therefore, FLDS provides opportunities for deep surveys of RdRp and the identification of novel genes that are distinct from the viral genes in public databases.

Function of microbial RNA viruses in aquatic ecosystems
Knowledge on the ecological functions of acute-type microbial RNA viruses is based on the effects of host cell lysis. For example, previous studies suggested the contribution of RNA viruses to the virus shunt in pelagic zones (Kaneko et al., 2021) and soil environments (Starr et al., 2019). In addition, a sequence match between a CRISPR spacer and a bacterial RNA virus was reported (Wolf et al., 2020). In contrast, their impacts on the evolution of microbial hosts have not yet been revealed. In the case of animals, RNA viral sequences incorporated into the host genome protect the host organism from related RNA viral infections (Suzuki et al., 2020). However, a similar system has not been reported for microbes infected with RNA viruses. The ability to mediate horizontal gene transfer, which is often reported for DNA viruses, has also not been reported.
In contrast, persistent-type microbial RNA viruses have a wide range of effects through the manipulation of their host organisms. For example, fungi with an RNA virus may induce thermal tolerance to plants, while those without the RNA virus do not (Marquez et al., 2007). Persistent-type RNA viruses also influence biological interactions between hosts and other organisms by changing secondary metabolites and pathogenicity (Chiba et al., 2009; Zhang et al., 2014; Aihara et al., 2018; Okada et al., 2018; Takahashi-Nakaguchi et al., 2019; Ninomiya et al., 2020). In some cases, persistent-type RNA viruses provide advantages to their hosts under specific conditions (Okada et al., 2018). Therefore, these viruses may affect the adaptation of microbes to environmental challenges (Bao and Roossinck, 2013). Many of these are examples in fungi, which serve as model systems for persistent-type RNA viral research; however, their functions in aquatic environments remain largely unknown. Nevertheless, the host-virus relationship and the impact of persistent-type RNA viral infection on the physiology of their hosts in terrestrial ecosystems imply that persistent-type RNA viruses in aquatic ecosystems also influence host physiology and subsequent ecological functions, including the niche adaptation of their host organisms.
Climate change due to global warming and ocean acidification, is an important issue for our planet. These environmental changes are expected to affect pelagic microbial ecosystems, and other impacts on marine microbial ecosystems and subsequent biogeochemical cycles may also accelerate environmental changes. Therefore, observations of the marine microbial community are essential for understanding microbial responses to a changing ocean. Furthermore, current updates in our understanding of the RNA virome suggest that acute- and persistent-type microbial RNA viruses both play a significant role in biogeochemical cycles via the viral shunt and the regulation of host physiology. Accordingly, we need to pay close attention to marine RNA viromes in the changing ocean even though pelagic RNA viromes have so far been overlooked in marine microbial ecology.
Citation
Urayama, S., Takaki, Y., Chiba, Y., Zhao, Y., Kuroki, M., Hagiwara, D., and Nunoura, T. (2022) Eukaryotic Microbial RNA Viruses—Acute or Persistent? Insights into Their Function in the Aquatic Ecosystem. Microbes Environ 37: ME22034.
https://doi.org/10.1264/jsme2.ME22034
Acknowledgements
This study was supported by a grant from the Institute for Fermentation, Osaka, by JSPS KAKENHI (Grant No. 18K19358, 20K20377, 20H05579, 21K18217, 21K20567, 22H04879, and 22J40078), and by Grants‐in‐Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Science, Sports, and Technology (MEXT) of Japan (Grant Nos. 16H06429, 16K21723, and 16H06437).
References
- Aihara, M., Urayama, S., Le, M.T., Katoh, Y., Higashiura, T., Fukuhara, T., et al. (2018) Infection by Magnaporthe oryzae chrysovirus 1 strain A triggers reduced virulence and pathogenic race conversion of its host fungus, Magnaporthe oryzae. J Gen Plant Pathol
84: 92–103.
- Banks, G.T., Buck, K.W., Chain, E.B., Darbyshire, J.E., and Himmelweit, F. (1969) Virus-like particles in penicillin producing strains of Penicillium chrysogenum. Nature
222: 89–90.
- Bao, X., and Roossinck, M.J. (2013) Multiplexed interactions: viruses of endophytic fungi. In Advances in Virus Research. Amsterdam: Elsevier, pp. 37–58.
- Beijerinck, M.W. (1898) Concerning a contagium vivum fluidum as cause of the spot disease of tobacco leaves. Phytopathol Classics
7: 33–52.
- Bian, R., Andika, I.B., Pang, T., Lian, Z., Wei, S., Niu, E., et al. (2020) Facilitative and synergistic interactions between fungal and plant viruses. Proc Natl Acad Sci U S A
117: 3779–3788.
- Botella, L., Janoušek, J., Maia, C., Jung, M.H., Raco, M., and Jung, T. (2020) Marine oomycetes of the genus Halophytophthora harbor viruses related to bunyaviruses. Front Microbiol
11: 1467.
- Breitbart, M., Salamon, P., Andresen, B., Mahaffy, J.M., Segall, A.M., Mead, D., et al. (2002) Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci U S A
99: 14250–14255.
- Brussaard, C.P., Noordeloos, A.A., Sandaa, R.A., Heldal, M., and Bratbak, G. (2004) Discovery of a dsRNA virus infecting the marine photosynthetic protist Micromonas pusilla. Virology
319: 280–291.
- Cai, G., Krychiw, J.F., Myers, K., Fry, W.E., and Hillman, B.I. (2013) A new virus from the plant pathogenic oomycete Phytophthora infestans with an 8 kb dsRNA genome: the sixth member of a proposed new virus genus. Virology
435: 341–349.
- Callanan, J., Stockdale, S.R., Shkoporov, A., Draper, L.A., Ross, R.P., and Hill, C. (2020) Expansion of known ssRNA phage genomes: from tens to over a thousand. Sci Adv
6: eaay5981.
- Chiba, S., Salaipeth, L., Lin, Y.H., Sasaki, A., Kanematsu, S., and Suzuki, N. (2009) A novel bipartite double-stranded RNA Mycovirus from the white root rot Fungus Rosellinia necatrix: molecular and biological characterization, taxonomic considerations, and potential for biological control. J Virol
83: 12801–12812.
- Chiba, Y., Tomaru, Y., Shimabukuro, H., Kimura, K., Hirai, M., Takaki, Y., et al. (2020) Viral RNA genomes identified from marine macroalgae and a diatom. Microbes Environ
35: ME20016.
- Culley, A. (2017) New insight into the RNA aquatic virosphere via viromics. Virus Res
244: 84–89.
- Culley, A.I., Lang, A.S., and Suttle, C.A. (2006) Metagenomic analysis of coastal RNA virus communities. Science
312: 1795–1798.
- Culley, A.I., Suttle, C.A., and Steward, G.F. (2010) Characterization of the diversity of marine RNA viruses. In Manual of Aquatic Viral Ecology. Wilheim, S., Weinbauer, M., and Suttle, C. (eds). Waco, TX: American Society of Limnology and Oceanography (ASLO), pp. 193–201.
- Culley, A.I., Mueller, J.A., Belcaid, M., Wood-Charlson, E.M., Poisson, G., and Steward, G.F. (2014) The characterization of RNA viruses in tropical seawater using targeted PCR and metagenomics. mBio
5: e01210–01214.
- Decker, C.J., and Parker, R. (2014) Analysis of double-stranded RNA from microbial communities identifies double-stranded RNA virus-like elements. Cell Rep
7: 898–906.
- Decker, C.J., Steiner, H.R., Hoon-Hanks, L.L., Morrison, J.H., Haist, K.C., Stabell, A.C., et al. (2019) dsRNA-Seq: identification of viral infection by purifying and sequencing dsRNA. Viruses
11: 943.
- Djikeng, A., Kuzmickas, R., Anderson, N.G., and Spiro, D.J. (2009) Metagenomic analysis of RNA viruses in a fresh water lake. PLoS One
4: e7264.
- Dolja, V.V., and Koonin, E.V. (2012) Capsid-less RNA viruses. eLS. URL https://doi.org/10.1002/9780470015902.a0023269
- Edgar, R.C., Taylor, J., Lin, V., Altman, T., Barbera, P., Meleshko, D., et al. (2022) Petabase-scale sequence alignment catalyses viral discovery. Nature
602: 142–147.
- Edwards, R.A., and Rohwer, F. (2005) Viral metagenomics. Nat Rev Microbiol
3: 504–510.
- Fukasawa, F., Hirai, M., Takaki, Y., Shimane, Y., Thomas, C.E., Urayama, S., et al. (2020) A new polycipivirus identified in Colobopsis shohki. Arch Virol
165: 761–763.
- Ghabrial, S.A., Castón, J.R., Jiang, D., Nibert, M.L., and Suzuki, N. (2015) 50-plus years of fungal viruses. Virology
479: 356–368.
- Ghosh, S., and Malik, Y.S. (2021) The true host/s of picobirnaviruses. Front Vet Sci
7: 615293.
- Goodman, R.P., Ghabrial, S.A., Fichorova, R.N., and Nibert, M.L. (2011) Trichomonasvirus: a new genus of protozoan viruses in the family Totiviridae. Arch Virol
156: 171–179.
- Hillman, B.I., and Cai, G. (2013) The family Narnaviridae: simplest of RNA viruses. Adv Virus Res
86: 149–176.
- Hirai, M., Takaki, Y., Kondo, F., Horie, M., Urayama, S., and Nunoura, T. (2021) RNA viral metagenome analysis of subnanogram dsRNA using fragmented and primer ligated dsRNA sequencing (FLDS). Microbes Environ
36: ME20152.
- Huang, Y., Niu, B., Gao, Y., Fu, L., and Li, W. (2010) CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics
26: 680–682.
- Izumi, T., Morioka, Y., Urayama, S., Motooka, D., Tamura, T., Kawagishi, T., et al. (2021) DsRNA sequencing for RNA virus surveillance using human clinical samples. Viruses
13: 1310.
- Kaneko, H., Blanc-Mathieu, R., Endo, H., Chaffron, S., Delmont, T.O., Gaia, M., et al. (2021) Eukaryotic virus composition can predict the efficiency of carbon export in the global ocean. iScience
24: 102002.
- Keel, B.N., Deng, B., and Moriyama, E.N. (2018) MOCASSIN-prot: a multi-objective clustering approach for protein similarity networks. Bioinformatics
34: 1270–1277.
- Khoshnan, A., and Alderete, J. (1994) Trichomonas vaginalis with a double-stranded RNA virus has upregulated levels of phenotypically variable immunogen mRNA. J Virol
68: 4035–4038.
- King, A.M.Q., Adams, M.J., Carstens, E.B., and Lefkowitz, E.J. (2012) Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. Amsterdam: Elsevier.
- Koonin, E.V., Dolja, V.V., and Krupovic, M. (2015) Origins and evolution of viruses of eukaryotes: The ultimate modularity. Virology
479–480: 2–25.
- Koonin, E.V., Dolja, V.V., Krupovic, M., Varsani, A., Wolf, Y.I., Yutin, N., et al. (2020) Global organization and proposed megataxonomy of the virus world. Microbiol Mol Biol Rev
84: e00061-00019.
- Kotta-Loizou, I., and Coutts, R.H. (2017) Mycoviruses in Aspergilli: a comprehensive review. Front Microbiol
8: 1699.
- Lachnit, T., Thomas, T., and Steinberg, P. (2016) Expanding our understanding of the seaweed holobiont: RNA viruses of the red alga Delisea pulchra. Front Microbiol
6: 1489.
- Lang, A.S., Culley, A.I., and Suttle, C.A. (2004) Genome sequence and characterization of a virus (HaRNAV) related to picorna-like viruses that infects the marine toxic bloom-forming alga Heterosigma akashiwo. Virology
320: 206–217.
- Lang, A.S., Vlok, M., Culley, A.I., Suttle, C.A., Takao, Y., Tomaru, Y., and ICTV Report Consortium (2021) ICTV virus taxonomy profile: Marnaviridae 2021. J Gen Virol
102: 001633.
- Liu, Y.C., Linder-Basso, D., Hillman, B.I., Kaneko, S., and Milgroom, M.G. (2003) Evidence for interspecies transmission of viruses in natural populations of filamentous fungi in the genus Cryphonectria. Mol Ecol
12: 1619–1628.
- Loeffler, F., and Frosch, P. (1898) Report of the commission for research on foot-and-mouth disease. Zent Bakt Parasitkde Abt I
23: 371–391.
- Marquez, L.M., Redman, R.S., Rodriguez, R.J., and Roossinck, M.J. (2007) A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science
315: 513–515.
- Márquez, L.M., and Roossinck, M.J. (2012) Do persistent RNA viruses fit the trade-off hypothesis of virulence evolution?
Curr Opin Virol
2: 556–560.
- Mihara, T., Nishimura, Y., Shimizu, Y., Nishiyama, H., Yoshikawa, G., Uehara, H., et al. (2016) Linking virus genomes with host taxonomy. Viruses
8: 66.
- Morris, T., and Dodds, J. (1979) Isolation and analysis of double-stranded RNA from virus-infected plant and fungal tissue. Phytopathology
69: 854–858.
- Nasir, A., Forterre, P., Kim, K.M., and Caetano-Anollés, G. (2014) The distribution and impact of viral lineages in domains of life. Front Microbiol
5: 194.
- Nerva, L., Ciuffo, M., Vallino, M., Margaria, P., Varese, G.C., Gnavi, G., and Turina, M. (2016) Multiple approaches for the detection and characterization of viral and plasmid symbionts from a collection of marine fungi. Virus Res
219: 22–38.
- Nibert, M.L., Ghabrial, S.A., Maiss, E., Lesker, T., Vainio, E.J., Jiang, D., and Suzuki, N. (2014) Taxonomic reorganization of family Partitiviridae and other recent progress in partitivirus research. Virus Res
188: 128–141.
- Ninomiya, A., Urayama, S., Suo, R., Itoi, S., Fuji, S.I., Moriyama, H., and Hagiwara, D. (2020) Mycovirus-induced tenuazonic acid production in a rice blast fungus Magnaporthe oryzae. Front Microbiol
11: 1641.
- Nuss, D.L. (2005) Hypovirulence: mycoviruses at the fungal-plant interface. Nat Rev Microbiol
3: 632–642.
- Obeng, N., Pratama, A.A., and Elsas, J.D.V. (2016) The significance of mutualistic phages for bacterial ecology and evolution. Trends Microbiol
24: 440–449.
- Okada, R., Ichinose, S., Takeshita, K., Urayama, S., Fukuhara, T., Komatsu, K., et al. (2018) Molecular characterization of a novel mycovirus in Alternaria alternata manifesting two-sided effects: Down-regulation of host growth and up-regulation of host plant pathogenicity. Virology
519: 23–32.
- Pratama, A.A., and van Elsas, J.D. (2018) The ‘neglected’ soil virome—Potential role and impact. Trends Microbiol
26: 649–662.
- Reed, W., and Carroll, J. (1901) The prevention of yellow fever. Public Health Pap Rep
27: 113–129.
- Roossinck, M.J. (2010) Lifestyles of plant viruses. Philos Trans R Soc Lond B Biol Sci
365: 1899–1905.
- Roossinck, M.J., Saha, P., Wiley, G.B., Quan, J., White, J.D., Lai, H., et al. (2010) Ecogenomics: using massively parallel pyrosequencing to understand virus ecology. Mol Ecol
19
Suppl 1: 81–88.
- Roossinck, M.J. (2011) The good viruses: viral mutualistic symbioses. Nat Rev Microbiol
9: 99–108.
- Roossinck, M.J. (2012) Persistent plant viruses: Molecular hitchhikers or epigenetic elements? In Viruses: Essential Agents of Life. Witzany, G. (ed.) Dordrecht: Springer Netherlands, pp. 177–186.
- Roossinck, M.J. (2015) Metagenomics of plant and fungal viruses reveals an abundance of persistent lifestyles. Front Microbiol
5: 767.
- Roossinck, M.J. (2019) Evolutionary and ecological links between plant and fungal viruses. New Phytol
221: 86–92.
- Sadeghi, M., Tomaru, Y., and Ahola, T. (2021) RNA viruses in aquatic unicellular eukaryotes. Viruses
13: 362.
- Schmitt, M.J., and Breinig, F. (2002) The viral killer system in yeast: from molecular biology to application. FEMS Microbiol Rev
26: 257–276.
- Shi, M., Lin, X.D., Tian, J.H., Chen, L.J., Chen, X., Li, C.X., et al. (2016) Redefining the invertebrate RNA virosphere. Nature
540: 539–543.
- Starr, E.P., Nuccio, E.E., Pett-Ridge, J., Banfield, J.F., and Firestone, M.K. (2019) Metatranscriptomic reconstruction reveals RNA viruses with the potential to shape carbon cycling in soil. Proc Natl Acad Sci U S A
116: 25900–25908.
- Steward, G.F., Culley, A.I., Mueller, J.A., Wood-Charlson, E.M., Belcaid, M., and Poisson, G. (2013) Are we missing half of the viruses in the ocean?
ISME J
7: 672–679.
- Stuart, K.D., Weeks, R., Guilbride, L., and Myler, P.J. (1992) Molecular organization of Leishmania RNA virus 1. Proc Natl Acad Sci U S A
89: 8596–8600.
- Suttle, C.A. (2016) Environmental microbiology: Viral diversity on the global stage. Nat Microbiol
1: 16205.
- Suzuki, Y., Baidaliuk, A., Miesen, P., Frangeul, L., Crist, A.B., Merkling, S.H., et al. (2020) Non-retroviral endogenous viral element limits cognate virus replication in Aedes aegypti ovaries. Curr Biol
30: 3495–3506.e6.
- Tai, V., Lawrence, J.E., Lang, A.S., Chan, A.M., Culley, A.I., and Suttle, C.A. (2003) Characterization of HaRNAV, a single‐stranded RNA virus causing lysis of Heterosigma akashiwo (raphidophyceae) 1. J Phycol
39: 343–352.
- Takahashi-Nakaguchi, A., Shishido, E., Yahara, M., Urayama, S., Sakai, K., Chibana, H., et al. (2019) Analysis of an intrinsic mycovirus associated with reduced virulence of the human pathogenic fungus Aspergillus fumigatus. Front Microbiol
10: 3045.
- Takao, Y., Nagasaki, K., Mise, K., Okuno, T., and Honda, D. (2005) Isolation and characterization of a novel single-stranded RNA virus infectious to a marine fungoid protist, Schizochytrium sp. (Thraustochytriaceae, Labyrinthulea). Appl Environ Microbiol
71: 4516–4522.
- Takao, Y., Mise, K., Nagasaki, K., Okuno, T., and Honda, D. (2006) Complete nucleotide sequence and genome organization of a single-stranded RNA virus infecting the marine fungoid protist Schizochytrium sp. J Gen Virol
87: 723–733.
- Tomaru, Y., Katanozaka, N., Nishida, K., Shirai, Y., Tarutani, K., Yamaguchi, M., and Nagasaki, K. (2004) Isolation and characterization of two distinct types of HcRNAV, a single-stranded RNA virus infecting the bivalve-killing microalga Heterocapsa circularisquama. Aquat Microb Ecol
34: 207–218.
- Torrecilhas, A.C., Soares, R.P., Schenkman, S., Fernandez-Prada, C., and Olivier, M. (2020) Extracellular vesicles in trypanosomatids: host cell communication. Front Cell Infect Microbiol
10: 602502.
- Uchida, K., Sakuta, K., Ito, A., Takahashi, Y., Katayama, Y., Omatsu, T., et al. (2021) Two novel endornaviruses co-infecting a phytophthora pathogen of Asparagus officinalis modulate the developmental stages and fungicide sensitivities of the host oomycete. Front Microbiol
12: 122.
- Uehara-Ichiki, T., Urayama, S., Hirai, M., Takaki, Y., Nunoura, T., Fujinaga, M., and Hanada, K. (2021) Complete genome sequence of Sikte (Sitke) waterborne virus, a member of the genus Tombusvirus. Arch Virol
166: 991–994.
- Urayama, S., Takaki, Y., and Nunoura, T. (2016) FLDS: A comprehensive dsRNA sequencing method for intracellular RNA virus surveillance. Microbes Environ
31: 33–40.
- Urayama, S., Takaki, Y., Nishi, S., Yoshida-Takashima, Y., Deguchi, S., Takai, K., and Nunoura, T. (2018a) Unveiling the RNA virosphere associated with marine microorganisms. Mol Ecol Resour
18: 1444–1455.
- Urayama, S., Takaki, Y., Nunoura, T., and Miyamoto, N. (2018b) Complete genome sequence of a novel RNA virus identified from a deep-sea animal, Osedax japonicus. Microbes Environ
33: 446–449.
- Urayama, S., Doi, N., Kondo, F., Chiba, Y., Takaki, Y., Hirai, M., et al. (2020a) Diverged and active partitiviruses in lichen. Front Microbiol
11: 561344.
- Urayama, S., Takaki, Y., Hagiwara, D., and Nunoura, T. (2020b) dsRNA-seq reveals novel RNA virus and virus-like putative complete genome sequences from Hymeniacidon sp. sponge. Microbes Environ
35: ME19132.
- Vainio, E.J., Hakanpää, J., Dai, Y.-C., Hansen, E., Korhonen, K., and Hantula, J. (2011) Species of Heterobasidion host a diverse pool of partitiviruses with global distribution and interspecies transmission. Fungal Biol
115: 1234–1243.
- van der Lende, T.R., Harmsen, M.C., and Wessels, J.G. (1994) Double-stranded RNAs and proteins associated with the 34 nm virus particles of the cultivated mushroom Agaricus bisporus. J Gen Virol
75: 2533–2536.
- Wang, A.L., and Wang, C.C. (1986) Discovery of a specific double-stranded RNA virus in Giardia lamblia. Mol Biochem Parasitol
21: 269–276.
- Wickner, R.B. (1996) Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol Rev
60: 250–265.
- Wilhelm, S.W., and Suttle, C.A. (1999) Viruses and nutrient cycles in the sea: viruses play critical roles in the structure and function of aquatic food webs. Bioscience
49: 781–788.
- Wolf, Y.I., Silas, S., Wang, Y., Wu, S., Bocek, M., Kazlauskas, D., et al. (2020) Doubling of the known set of RNA viruses by metagenomic analysis of an aquatic virome. Nat Microbiol
5: 1262–1270.
- Xu, P., Chen, F., Mannas, J.P., Feldman, T., Sumner, L.W., and Roossinck, M.J. (2008) Virus infection improves drought tolerance. New Phytol
180: 911–921.
- Zayed, A.A., Wainaina, J.M., Dominguez-Huerta, G., Pelletier, E., Guo, J., Mohssen, M., et al. (2022) Cryptic and abundant marine viruses at the evolutionary origins of Earth’s RNA virome. Science
376: 156–162.
- Zhang, R., Liu, S., Chiba, S., Kondo, H., Kanematsu, S., and Suzuki, N. (2014) A novel single-stranded RNA virus isolated from a phytopathogenic filamentous fungus, Rosellinia necatrix, with similarity to hypo-like viruses. Front Microbiol
5: 360.
- Zimmerman, A.E., Howard-Varona, C., Needham, D.M., John, S.G., Worden, A.Z., Sullivan, M.B., et al. (2020) Metabolic and biogeochemical consequences of viral infection in aquatic ecosystems. Nat Rev Microbiol
18: 21–34.