Abstract
Due to the high crude fiber content, straw of various crops is difficult to become a high quality forage resource. The degradation of cellulose in nature mainly depends on the cellulase secreted by microbes, which degrade cellulose into small molecular substances through chemical action, and the microbes that secrete cellulase mainly include some bacteria, fungi and actinomycetes, etc. The large and diverse microbial population contained in the mammalian gastrointestinal tract plays an important role in nutrient digestion. At present, many cellulose-degrading strains have been screened and obtained from animal digestive system and feces, such as Bacillus subtilis from the feces of Panda, Bacillus amyloliquefaciens from the cecum of goose. In this study, the fungal diversity was analysed in the fresh faeces of Tibetan sheep, Tibetan gazelle and Tibetan antelope in Qiangtang, Tibet. Results showed that the structure and species of gut fungi are different in three animals, which may be related to the different physiological functions among different animals, e.g., Tibetan antelope and Tibetan gazelle have stronger tolerance to rough feeding than Tibetan sheep. This study will lay a foundation for cellulose-degrading fungal development and provides technical support for improving rough feeding tolerance of Tibetan sheep.
Introduction
The sheep industry has a long history in Tibet and is an important pillar for development of agriculture and husbandry. In the past decade, the number of Tibetan sheep in Tibet has remained at about 1.00×107, accounting for 1/2 of the total number of livestock in Tibet and ranking first among other livestock (Liu, Ren, Dawa, Ciwang, & Ni, 2018). However, the high crude fiber content of forage and the shortage of high quality forage are the important factors that impede the development of the Tibetan sheep industry. At present, the best way to utilize cellulose is through microbial degradation (Liu, Zhang, Yang, & Ju, 2017) and as the main digestive organ of animals, the intestine contains a variety of functional microorganisms. Therefore, it is of great significance to analyze the intestinal microbes of animals to develop functional strains.
As a new type of functional microorganisms, animal faeces can produce enzyme preparations, antifungal agents, antibacterial agents and other products (Weber & Gloer, 1991; Soman, Gloer, Koster, & Malloch, 1999), the function and potential application of fecal microbes have gradually received increasing attention (Sampson & Mazmanian, 2015; Dinan & Cryan, 2012). Numerous microbes can parasitise the intestinal tract of herbivores and provide the animal with energy and nutrients by degrading indigestible fibres, such as cellulose, hemicellulose and lignin (Mamun et al., 2019; Husso et al., 2020). Most microbes in animal intestines are bacteria, but some fungi also exist and the studies of fungi are limited compared to bacteria (Li et al., 2017; Yang et al., 2017). Animal fungi have a strong ability to degrade crude fibres and can effectively improve the utilisation rate of roughage by animals (Hagen et al., 2021). Study of fungi in animal faeces has great significance for development of new microbial sources. Researchers have identified the following six fungal genera that can degrade plant cell walls, Neocallimastix, Piromyces, Orpinomyces, Anaeromyces, Caecomyces and Cyllamyces, respectively (James et al., 2006; Chen et al., 2007). High levels of diversity and community structure of gut microbiota are strongly associated with host species and are very stable and consistent within host species. In addition, the microbial diversity index is an objective indicator to analyze the richness and evenness of Microbial flora, so the determination of fecal fungal diversity and structure are helpful to analyze the species and quantity distribution of gut microbiota (Rawls, Mahowald, Ley, & Gordon, 2006; Wei, Morrison, & Yu, 2013).
Faecal microbial transplantation technology can significantly change the structure of the intestinal flora of animals, the intestinal physiological characteristics and physiological indicators (such as intestinal index, morphology and digestive enzymes) become similar to those of donor animals, thereby improving their physiological function (Diao et al., 2016). Tibetan antelope, Tibetan gazelle and Tibetan sheep belong to Caprinae; As wild animals, Tibetan antelope and Tibetan gazelle have high coarse feeding tolerance and strong disease resistance which maybe related to the unique microbial community structures (Shi, Miao, Su, & Wasser, 2021). So the present study examines the faeces fungi community of Caprinae animals based on the potential of transplantation of fecal microbes and enhances the cellulose degradation capacity by screening cellulose degrading fungal. Results can provide a theoretical basis for screening functional superior strains and lay a foundation for faecal transplantation of Caprinae.
Materials and methods
Ethics statement
This study was approved and instructed by the Animal Collaborative Research Center of Tibet Agriculture and Animal Husbandry College, Tibet, China (Unified social credit code:12540000MB0P013721).
Research location and sample collection
In this study, the Tibetan sheep (Ovis aries), Tibetan gazelle (Procapra picticaudata) and Tibetan antelope (Pantholops hodgsonii) in Qiangtang Nature Reserve of Tibet were investigated. Faecal samples of the three animals were collected from areas surrounding Qiangtang National Nature Reserve. The vegetation type is alpine steppe and mainly includes zonal vegetation, such as Stipa pubescens in this region. The area has an average elevation of over 5000 masl. The climate is cold and dry, air is thin and the natural environment is harsh. The temperature widely changes throughout the day. The average annual temperature is close to -10 °C and the annual precipitation is about 150 mm, of which over 80% is concentrated from Jun to Sep (Hu, Du, & Yuan, 2019).
In this study, Tibetan sheep, Tibetan antelope and Tibetan gazelle were all raised by grazing without artificial supplementary feeding. A group of Tibetan sheep (approximately 120) was found in the grasslands near Shuanghu County (33°30´N, 88°83´E, elevation: 5,010 masl); Two groups of Tibetan antelope (approximately 10) were found at two sites in the reserve (32°73´N, 88°91´E, elevation: 4,969 masl; 32°64´N, 88°92´E, elevation: 4,878 masl). Two groups of Tibetan gazelle (approximately 9) were observed at two sites (31.39´N, 90.35´E, elevation: 4,641 masl; 32°54´N, 88°93´E, elevation: 4,843 masl). The telescope was used to track the areas where the animals defecated. Five fresh faecal samples were randomly selected for each animal after they had left the area (In order to avoid collecting the feces from the same animal individuals, we collected feces were >50 m apart from each other). The outer layer of the faeces was removed using sterilised toothpicks, and uncontaminated fresh faeces were collected and placed in 20 mL cryopreservation tubes. The samples were stored at −20 °C and subsequently analysed to determine their fungal microbial diversity.
Total DNA Extraction
Fungal genomic DNA was extracted by using QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) following the producer's instructions. DNA quality was determined by electrophoresis on agarose gel 0.8% (w/v) and the quantity was determined by using NanodropTMspectrophotometer (NanoDrop Technologies, Thermo Scientific, USA).
ITS hypervariable region gene amplification
Specific PCR primers (ITS5F: GGA AGT AAA AGT CGT AAC AAG G and ITS2R: GCT GCG TTC TTC ATC GAT GC) based on conserved regions were synthesized to amplify the ITS1 regions. AxyPrep DNA Gel Extraction Kit (Axygen, CA, USA) and the 2% agarose gel electrophoresis were used for target fragment recovery and PCR amplification product evaluation, respectively. The PCR amplification recovery products were quantified on fluorescent Microplate reader (BioTek, FLx800) via using Quant-iT PicoGreen dsDNA Assay Kit based on the preliminary quantitative results of electrophoresis. For sequence library construction, TruSeq Nano DNA Low Throughput Library Prep Kit (Illumina, CA, USA) was used following manufacturer's specifications. Amplified products' sequence ends were repaired by End Repair Mix2. To enrich the sequencing library template, PCR amplification was carried out and the library enrichment product was purified again via using BECKMAN AMPure XP Beads. The final fragment-selection and purification of library was performed by using 2% agarose gel electrophoresis.
The quality of libraries was examined on Agilent Bioanalyzer using Agilent High Sensitivity DNA Kit prior to sequencing procedure. The libraries with only one peak signal and no linker signal were considered for the procedure. Moreover, the libraries were quantified using Quant-iT PicoGreen dsDNA Assay Kit on Promega QuantiFluor fluorescence quantification system. The qualified library concentration must be above 2 nM. The qualified libraries were gradient diluted and mixed in proportion according to the amount of sequencing required. The MiSeq Reagent Kit V3 (600 cycles) was used to perform 2×300 bp paired-end sequencing on the MiSeq sequencing machine after the mixed libraries were denatured into single strands by sodium hydroxide.
Bioinformatics and Statistical Analysis
The QIIME software (Qiime1.9.1) was used for ITS rRNA original data quality preliminary screening and analysis. Interrogative and short sequences (<200 bp) were discarded using QIIME software. The obtained sequences were clustered and operational taxonomic unit (OTU) partitioned at ≥97% sequence similarity by using clustering program VSEARCH (1.9.6.). Representative sequence of each OTU was classified and grouped by comparing with those in the Unite database (Release 7.0, https://unite.ut.ee/) (Kõljalg et al., 2013). Additionally, the obtained sequences with 97% similarity were merged to the same operational taxonomic units (OTUs). Before calculating alpha and beta diversity statistics, sequencing depth of each sample was evaluated via using sparse curves. Continuous analysis of alpha diversity and beta diversity were performed based on the output normalized date. Four metrics including Chao1, Abundance-based coverage estimator (ACE), Simpson and Shannon were used to analyze alpha diversity by IBM SPSS (22.0) statistics software. Beta diversity was analyzed by utilizing the similarity of microbial community structures among different groups through principal component analysis (PCA) (Ramette, 2007). The Metastats statistical algorithm was used to analyze the discrepancy of the microbial communities between groups at phylum and genus level (White, Nagarajan, & Pop, 2009). R (v3.0.3) software were used for statistical analysis (Liu et al., 2020). The criterion of significance was performed at p-values less than 0.05 and the values were presented as means ± SD.
Result
Sequencing results and OTU cluster statistical analysis
A total of 157611, 201270 and 168822 high-quality valid sequences were obtained from the Tibetan sheep, Tibetan gazelle and Tibetan antelope, respectively. The average length of these sequences was approximately 100-350 bp (Fig. 1). The shannon and Chao1 rarefaction curves demonstrated that the sequence numbers, abundance and evenness of fungal species in the sample meet the requirement for sequencing and analysis (Fig. 2). The sequences were clustered into 797 OTUs by 97% sequence identity. A total of 374, 388 and 293 OTUs were identified in Tibetan sheep, Tibetan gazelle and Tibetan antelope, respectively. 59 OTUs of which were common among the three animals (Fig. 3). Interestingly, 211 fungal species found in Tibetan sheep were not found in Tibetan antelope or Tibetan gazelle. Moreover, 257 fungal species found in Tibetan gazelle and 130 fungal species found in Tibetan antelope were not found in the other animals. The sequence data has been deposited at GenBank under the accession no. PRJNA821987.

Fig. 1 - Sequencing statistical analysis results in different animals. A: Statistics of effective sequences in each sample. B: The length distribution of sequences of samples from Tibetan sheep, Tibetan gazelle and Tibetan antelope.

Fig. 2 - The diversity index rarefaction curve of shannon and Chao1 of each samples.

Fig. 3 - Venn map of comparison of > OTUs distribution in three animals.
The Chao1 and ACE indices were 137.58 and 143.64, 121.51 and 121.88, 77.38 and 79.58 in Tibetan sheep, Tibetan gazelle and Tibetan antelope, respectively, with no significant difference among different groups (P > 0.05). Chao1 and ACE indexes demonstrated no visible differences in flora evenness among the three animals (Fig. 4). Simpson index was 0.96, 0.88 and 0.60 in Tibetan sheep, Tibetan gazelle and Tibetan antelope, respectively, Tibetan sheep and Tibetan gazelle were obviously higher than Tibetan antelope (P < 0.05). Shannon index was 5.45, 4.64 and 2.67 in Tibetan sheep, Tibetan gazelle and Tibetan antelope, respectively, Tibetan antelope was evident lower than Tibetan sheep and Tibetan gazelle (P < 0.05). Simpson and Shannon indexes revealed striking differences in flora evenness among the three animals (Fig. 4). However, obvious differences were watched in the principal component of microbial community structure in different animal groups by PCA analysis, especially between Tibetan antelope groups and Tibetan gazelle groups (Fig. 5). To further investigate the difference of faecal microbial composition among the different animals, ADONIS of weighted UniFrac distances indicated that there was a significant difference between the three animals (R2 = 0.5840, P = 0.001).

Fig. 4 - Comparison the alpha diversity index among different animals. A: Simpson index difference analysis; B: Shannon index difference analysis; C: Chao1 index difference analysis; D: ACE index difference analysis (**P < 0.01; No * indicate no significant difference, P > 0.05).
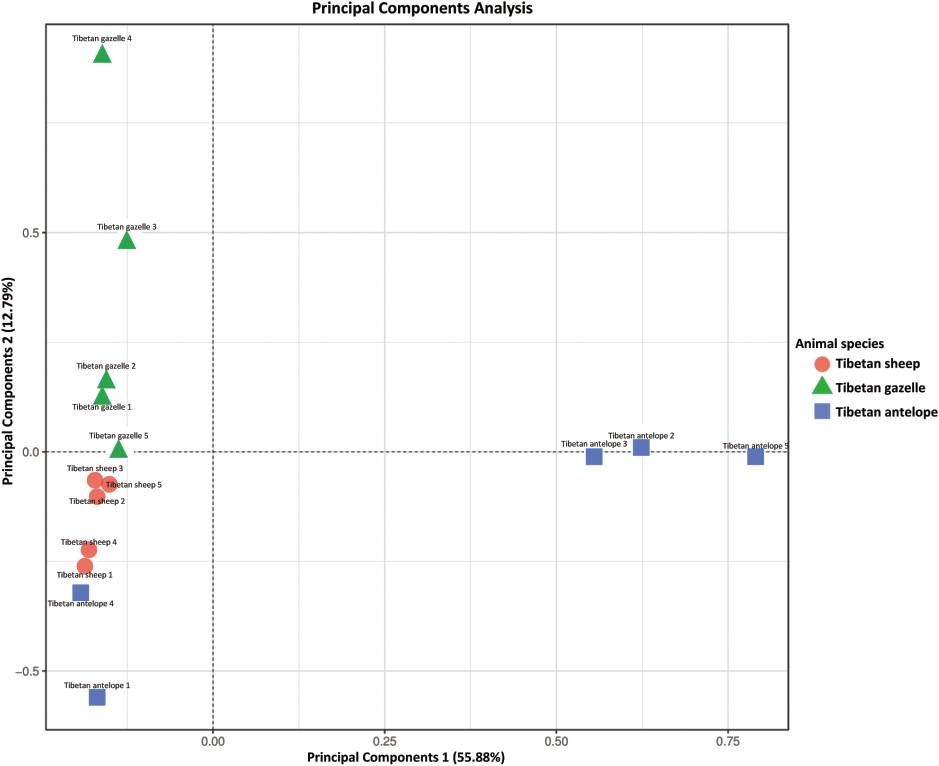
Fig. 5 - Principal Component analysis of the similarity of microbial community structure in different animals.
At the phylum level (Fig. 6A), Ascomycota and Basidiomycota contributed to most of the microbial communities in Tibetan sheep, Tibetan gazelle and Tibetan antelope. The relative abundance of Ascomycota (69.56%, 74.94% and 66.24% in three animals, respectively) in Tibetan antelope were lower than that in Tibetan sheep and Tibetan gazelle, Basidiomycota (25.16%, 21.56% and 32.68% in three animals, respectively) was higher than that in Tibetan sheep and Tibetan gazelle, while there were not significantly difference among the three animals (P > 0.05). Other phyla including Mortierellomycota, Neocallimastigomycota, Rozellomycota, Olpidiomycota, Mucoromycota and Glomeromycota were represented with a lower abundance accounting for less than 1% of the total ITS gene sequences in all samples. At the family level, the main families in the three animals were different. The most abundant families were Cordycipitaceae (11.78%), Ustilaginaceae (7.44%), Trichosporonacea (4.36%) and Plectosphaerellaceae (5.04%) in Tibetan sheep, Sporidiobolaceae (15.24%), Didymellaceae (13.50%), Stachybotryaceae (11.88%) and Thelebolaceae (2.80%) contributed to most abundant families in Tibetan gazelle, while Thelebolaceae (46.76%), Anthracoideaceae (11.44%), Filobasidiaceae (6.18%) and Ustilaginaceae (4.00%) were the most abundant families in the Tibetan antelope (Fig. 6B). In addition, the Thelebolus (16.79%), Sporobolomyces (5.44%), Didymella (4.58%), Lecanicillium (4.34%), Anthracoidea (4.28%), Ustilago (3.78%), Candida (3.69%), Naganishia (3.64%), Cutaneotrichosporon (2.17%), Sphenorchidium (1.64%) and Simplicillium (1.48%) were the predominant fungus genera in all sample (Fig. 6C, D). The comparison results of faecal microbiota among Tibetan sheep, Tibetan gazelle and Tibetan antelope by using Metastats showed that all fungus phyla in Tibetan sheep, Tibetan gazelle and Tibetan antelope were no significantly difference (P > 0.05). At the genus level (Fig. 7), 11 genera (including Verticillium, Lecanicillium, Phaeoacremonium, Cutaneotrichosporon, Malassezia, Urocystis, Ustilago, Antennariella, Didymella, Simplicillium
and Chaetomium) showed significant difference with p < 0.01 between Tibetan sheep and Tibetan gazelle, and 14 genera (including Myrothecium, Plenodomus, Thelebolus, Plectosphaerella, Neostagonospora and so on) were found significantly different with p < 0.05 between Tibetan sheep and Tibetan gazelle; Moreover, a comparison of Tibetan sheep and Tibetan antelope, we found 9 genera (including Verticillium, Urocystis, Phaeoacremonium, Simplicillium, Lecanicillium, Malassezia, Thelebolus, Chaetomium and Cutaneotrichosporon) showed significant difference with p < 0.01 and 7 genera (including Acremonium, Sporobolomyces, Cladosporium and so on) were found significantly different with p < 0.05

Fig. 6 - Relative abundance of gut fungal taxa at the different level. A: at the Phylum level; B: Family level; C: Genus level; D: Between group taxa assignments at Genus level.

Fig. 7 - Comparison of microbial community structure in different animals with Genus level by Metastats.
Discussion
In the Qinghai Tibet plateau areas, where herdsmen highly dependent on Tibetan sheep and Tibetan sheep products (Cui et al., 2019). Due to the higher crude fiber of forage, the shortage of high-quality forage has seriously limited the Tibetan sheep industry. Therefore, it is very meaningful to dig the cellulose degradation fungi of Tibetan sheep.
In this study, we obtained 89, 145 and 84 genera from the faeces of Tibetan sheep, Tibetan gazelle and Tibetan antelope, respectively. Beta diversity analysis showed that Tibetan sheep, Tibetan gazelle and Tibetan antelope gathered in different areas, indicating the different structures of fungal flora among different Caprinae animals. Alpha diversity analysis obtained that the fungal diversity of faeces in Tibetan sheep (captivity animal) were higher than those in Tibetan gazelle and Tibetan antelope (wild animal). This finding is inconsistent with previous reports on the gut bacterial composition between wild and captive Tibetan wild ass. This research obtained that the bacterial diversity of gut microbes in the wild Tibetan wild ass population is significantly higher than for those individuals in captivity (Gao et al., 2019). This may be related to three factors: First, the fungal diversity is different from the bacterial diversity; Second, the digestion methods of ruminants and monogastric herbivores were different. Moreover, both the feeding method and animal species were the main reason for the different structure of the intestinal flora in the animals (Yang, Bian, Su, & Zhu, 2014; Tian, Wu, Chen, Yu, & He, 2017). In this study, the feeding methods of Tibetan sheep, Tibetan antelope and Tibetan gazelle were consistent (grazing), and the animal species were different, but in Gao's research, the animal species were the same and the feeding methods were different (domestication and grazing). In the present study, Thelebolus, Sporobolomyces, Didymella, Lecanicillium, Anthracoidea, Ustilago, Candida, Naganishia, Cutaneotrichosporon, Sphenorchidium and Simplicillium were the dominant fungal communities in the three animals, inconsistent with the findings of urban soils fungi from China (Zhang et al., 2021), animal skin fungi from Italy (Allizond et al., 2016), mammalian herbivores faecal fungi from Puerto Rico (Richardson, 2008) and waterborne fungi from Africa (Magwaza, Nxumalo, Mamba, & Msagati, 2017). In the other four places, these listed genera were not dominant fungal, the discrepancy may be due to the habitat environment that can affect the fungal flora structure and the special low-oxygen environment in the plateau of the Qiangtang National Nature Reserve in Tibet is different from other places, whether these listed fungi prefer the low-oxygen environment need to be further studied. Previous sequencing results showed that Ascomycota and
Basidiomycota were the dominant species in the gastrointestinal track of humans and other animals (Li et al., 2018). In the present work, Ascomycota and Basidiomycota were the main phyla in Tibetan sheep, Tibetan gazelle and Tibetan antelope, but no significant difference was found among the three animals.
The intestinal flora is closely related to the nutrition, metabolism and immunity of the host (Chen, Helm, Gabler, Hostetter, & Burrough, 2020). Tibetan gazelle and Tibetan antelope live in an alpine environment with low vegetation coverage and long dry grass period, but they exhibit higher dry matter digestibility than other ruminants (Cao, Xu, Zhao, & Yu, 2007). Hence, Tibetan gazelle and Tibetan antelope have strong crude fibre digestion ability. Neocallimastix, Piromyces, Orpinomyces, Anaeromyces, Caecomyces and Cyllamyces have been identified can degrade crude fibre, but in our study, none of the above fungal genera were identified. Therefore, we inferred that some unreported fungi may have the function of degrading crude fibre in Caprinae animals. The number of Thelebolus in Tibetan gazelle and Tibetan antelope is significantly higher than that in Tibetan sheep. In addition, the abundance of Didymella, Anthracoidea, Neostagonospora, Plectosphaerella and Myrothecium in Tibetan gazelle are significantly higher than those in Tibetan sheep. Thelebolus is a filamentous Ascomycetous fungus that is isolated from plants in psychrophilic Antarctic; this fungus may have a role in adapting to low Antarctic temperatures by modulating membrane fluidity (Shiv, Paras, Sanjay, & Prabhat, 2013). Didymella is the only genus that is correctly associated with Phoma and Ascochyta; this genus is mainly identified in the stems and leaves of plants (McPartland, 2018), but related research remains limited. The main classification standard of Anthracoidea is the morphological characteristics of spores, including the size of spores and the external protrusion and internal expansion of the cell wall (He, Li, Chang, & Wang, 2011). This genus includes more than 80 species, and all of which are parasitic on Cyperaceae plants. Neostagonospora is identified from Rosaceae (Rose) (Wanasingle et al., 2018). Plectosphaerella is the largest genus in the family Plectosphaerellaceae. Some species are plant pathogens, whereas others are soil borne (Zhang et al., 2019). Myrothecium is an important biological resources for new and bioactive natural products which screened from the salt-resistant plant Apocynum venetum, due to Myrothecium can produce sesquiterpenes,
diterpenes and cyclopeptides (Xu et al., 2018). The six fungal genera can be identified in plants, but their specific physiological functions and their ability to degrade crude fibre in animal intestines remain to be further studied. Therefore, we suggest that cellulose degradation test should be conducted in vitro to explore whether these fungi have the function of degrading cellulose, or using third-generation sequencing technology to identify the fecal fungi species of three Caprinae animals, to further explore the relationship between these fungi and cellulose degradation. Transplanting the faeces of Tibetan antelope and Tibetan gazelle into Tibetan sheep should also be performed to explore whether it can improve the degradation rate of dry matter of Tibetan sheep.
Lecanicillium is an important biocontrol fungus which has the ability to reduce harmful fungi (Gurulingappa, McGee, & Sword, 2011); The metabolites of Simplicillium have a strong inhibition effect of gram-positive bacteria, such as Staphylococcus aureus, Bacillus subtilis and Micrococcus luteus (Chen, Huang, Li, & Tsay, 2008; Dong, Lin, Xing, Chen, & Han, 2014). In our study, the abundance of Lecanicillium and Simplicillium in Tibetan sheep are higher than those in Tibetan gazelle and Tibetan antelope. Interesting, the numbers of pathogenic fungi, such as Verticillium (Iulian et al., 2020), Urocystis (Kashyap et al., 2020), Phaeoacremonium (Horace, Arnaud, & Georgios, 2020), Malassezia (Malinovsk, Conkova, Vaczi, Harcarova, & Bohmova, 2019) and Cutaneotrichosporon (Reema, Pallabi, Biswajyoti, & Pratap, 2018) have the same changing trend in three animals. In general, beneficial fungi can compete with pathogenic fungi for colonization loci, hence, the function of the above beneficial fungi and pathogenic fungi need to be further studied in Tibetan sheep.
The faecal fungal flora structure was studied in Tibetan sheep, Tibetan gazelle and Tibetan antelope. The structure significantly differed among the three animals. This study has some limitations, such as several incontrollable important variables including individual variation and individual dietary habit. This work reveals the complexity of the faecal fungal flora of Caprinae in the Qinghai-Tibet Plateau, and its composition and function are of great importance to screen functional fungi and transplant animal faeces.
Disclosure
The authors declare no conflicts of interest. All the experiments undertaken in this study comply with the current laws of the country where it was performed.
Author contributions
QH Kong, SZ Liu, HH Wang and ZD Shang contributed to the initial design of this project. QH Kong, ZK Tan, P Shang and ZD Shang collected samples from different animals. ZD Shang and QH Kong conducted the experiment, conducted bioinformatics analyses and prepared the manuscript of this publication.
Acknowledgments
This study was supported by Tibet Regional Science and Technology Collaborative Innovation Project (QYXTZX-NQ2021-01), the Central Government supports the special fund project for the reform and development of local universities (KY2022ZY-01) and Tibetan Plateau Feed Processing Research Center Project (XZJYT2018GCZX).
References
- Allizond, V., Tullio, V., Cuffini, A.M., Roana, J., Scalas, D., Marra, E.S., Piersigilli, G., Merlino, C., Mandras, N., & Banche, G. (2016). Advances in Microbiology, Infectious Diseases and Public Health: Fungal Occurrence in the Hair and Skin of Symptomatic Pets in Turin, Italy. Advances in Experimental Medicine and Biology, 897, 55-62. https://doi.org/10.1007/5584_2015_5004
- Cao, J.H., Xu, S.X., Zhao, X.Q., & Yu, M.S. (2007). Herbage utilization of Tibetan antilope (Pantholops hodgsoni) during cold season. Acta Theriologica Sinica, 27(2), 206-208 (in China). https://doi.org/10.16829/j.slxb.2007.02.019
- Chen, R.S., Huang, C.C., Li, J.C., & Tsay, J.G. (2008). First report of Simplicillium lanosoniveum causing brown spot on Salvinia auriculata and S.molesta in Taiwan. Plant Disease, 92(11), 1589. https://doi.org/10.1094/PDIS-92-11-1589C
- Chen, Y.C., Ysai, S.D., Cheng, H.L., Chien, C.Y., Hu, C.Y., & Cheng, T.Y. (2007). Caecomyces sympodialis sp. nov., a new rumen fungus isolated from Bos indicus. Mycologia, 99(1), 125-130. https://doi.org/10.3852/mycologia.99.1.125
- Chen, Y.M., Helm, E.T., Gabler, N., Hostetter J.M., & Burrough, E.R. (2020). Alterations in Intestinal Innate Mucosal Immunity of Weaned Pigs During Porcine Epidemic Diarrhea Virus Infection. Veterinary pathology, 57(5), 642-652. https://doi.org/10.1177/0300985820932140
- Cui, X.X., Wang, Z.F., Yan, T.H., Chang, S.H., Wang, H., & Hou, F.J. (2019). Rumen bacterial diversity of Tibetan sheep (Ovis aries) associated with different forage types on the Qinghai-Tibetan Plateau. Canadian Journal of Microbiology, 65(12), 859-869. https://doi.org/10.1139/cjm-2019-0154
- Diao, H., Yan, H.L., Xiao, Y., Yu, B., Yu, J., He, J., Zheng, P., Zeng, B.H., Wei, H., Mao, X.B., & Chen, D.W. (2016). Intestinal microbiota could transfer host Gut characteristics from pigs to mice. BMC microbiology, 16, 238-253. https://doi.org/10.1186/s12866-016-0851-z
- Dinan, T.G., & Cryan, J.F. (2012). Regulation of the stress response by the gut microbiota: implications for psychoneuroendocrinology. Psychoneuroendocrinology, 37(9), 1369-1378. https://doi.org/10.1016/j.psyneuen.2012.03.007
- Dong, Q.L., Lin, T.Y., Xing, X.Y., Chen, B., & Han, Y. (2014). Identification of a symbiotic fungus from blue-green alga and its extracellular polysaccharide. Letters in Applied Microbiology, 58(4), 303-310. https://doi.org/10.1111/lam.12192
- Gao, H., Chi, X., Qin, W., Wang, L., Song, P., Cai, Z., Zhang, J., & Zhang, T. (2019). Comparison of the gut microbiota composition between the wild and captive Tibetan wild ass (Equus kiang). Journal of Applied Microbiology, 126(6), 1869-1878. https://doi.org/10.1111/jam.14240
- Gurulingappa, P., McGee, P.A., & Sword, G. (2011). Endophytic Lecanicillium lecanii and Beauveria bassiana reduce the survival and fecundity of Aphis gossypii following contact with conidia and secondarymetabolites. Crop Protection, 30(3), 349-353. https://doi.org/10.1016/j.cropro.2010.11.017
- Hagen, L.H., Brooke, C.G., Shaw, C.A., Norbeck, A.D., Piao, H., Arntzen, M., Olson, H.M., Copeland, A., Isern, N., Shukla, A., Roux, S., Lombard, V., Henrissat, B., O'Malley, M.A., Grigoriev, I.V., Tringe, S.G., Mackie, R.I., Pasa-Tolic, L., Pope, P.B., & Hess, M. (2021). Proteome specialization of anaerobic fungi during ruminal degradation of recalcitrant plant fiber. The ISME Journal, 15(2), 421-434. https://doi.org/10.1038/s41396-020-00769-x
- He, S.Q., Li, H.X., Chang, X.Y., & Wang, S.R. (2011). Anthracoidea maquensis, a new species of smut fungi from China. Mycological Progress, 10, 53-55. https://doi.org/10.1007/s11557-010-0672-7
- Horace, M., Arnaud, R., & Georgios, D.P. (2020). First report of a new corneal pathogen: Phaeoacremonium parasiticum. European Journal of Clinical Microbiology & Infectious Diseases, 39, 2477-2480. https://doi.org/10.1007/s10096-020-03980-y
- Husso, A., Jalanka, J., Alipour, M.J., Huhti, P., Kareskoski, M., Pessa-Morikawa, T., Iivanainen, A., & Niku, M. (2020). The composition of the perinatal intestinal microbiota in horse. Scientific reports, 10, 441-452. https://doi.org/10.1038/s41598-019-57003-8
- Hu, J., Du, J., & Yuan, L. (2019). Analysis on Variation Feature of Pan Evaporation and Effect Factors in Chang Tang Nature Reserve of Tibet during 1971-2017. Climate Change Research Letters, 8(3), 356-364 (in China). https://doi.org/10.12677/CCRL.2019.83040
- Iulian, G., Harmeet, S.C., Daniel, T.L., Tiedemann, A.V., Snowdon, R.J., & Obermeier, C. (2020). Gene presence-absence variation associates with quantitative Verticillium longisporum disease resistance in Brassica napus. Scientific Reports, 10(1), 2294-2316. https://doi.org/10.1038/s41598-020-61228-3
- James, T.Y., Letcher, P.M., Longcore, J.E., Mozley-Standridge, S.E., Porter, D., Powell, M.J., Griffith, G.W., & Vilgalys, R. (2006). A molecular phylogeny of the flagellated fungi (Chytridiomycota) and description of a new phylum (Blastocladiomycota). Mycologia, 98(6), 860-871. https://doi.org/10.3852/mycologia.98.6.860
- Kashyap, P.L., Kumar, S., Kumar, R.S., Sharma, A., Jasrotia, P., Singh, D.P., & Singh, G.P. (2020). Molecular Diagnostic Assay for Rapid Detection of Flag Smut Fungus (Urocystis agropyri) in Wheat Plants and Field Soil. Frontiers in Plant Science, 11, 1039. https://doi.org/10.3389/fpls.2020.01039
- Kõljalg, U., Nilsson, R.H., Abarenkov, K., Tedersoo, L., Taylor, A.F.S., Bahram, M., Bates, S. T., Bruns, T.D., Bengtsson-Palme, J., Callaghan, T.M., Douglas, B., Drenkhan, T., Eberhardt, U., Dueñas, M., Grebenc, T., Griffith, G.W., Hartmann, M., Kirk, P.M., Kohout, P., … Larsson, K.H. (2013). Towards a unified paradigm for sequence-based identification of fungi. Molecular Ecology, 22, 5271-5277. https://doi.org/10.1111/mec.12481
- Li, J., Chen, D., Yu, B., He, J., Zheng, P., Mao, X., Yu, J., Luo, J., Tian, G., Huang, Z., & Luo, Y. (2018). Fungi in gastrointestinal tracts of human and mice: from community to functions. Microbial Ecology, 75 (4), 821-829. https://doi.org/10.1007/s00248-017-1105-9
- Li, P., Niu, Q., Wei, Q., Zhang, Y., Ma, X., Kim, S. W., Lin, M., & Huang, R. (2017). Microbial shifts in the porcine distal gut in response to diets supplemented with Enterococcus Faecalis as alternatives to antibiotics. Scientific Reports, 7, 41395-41404. https://doi.org/10.1038/srep41395
- Liu, L., Zhang, S.Q., Yang, X., & Ju, M.J. (2017). Cellulose isolation from corn stalk treated by alkaline biochars in solvent systems. BioResources, 13(1), 691-703. https://doi.org/10.15376/biores.13.1.691-703
- Liu, M.J., Ren, Y., Dawa, D.Z., Ciwang, Y.Z., & Ni, Z. (2018). Development Status, Existing Problems and Corresponding Countermeasures of the Tibetan Mutton Sheep Industry. Animal Husbandry and Feed Science, 39(1), 60-68 (in china). http://doi.org/10.16003/j.cnki.issn1672-5190.2018.01.016
- Liu, Z., Li, A., Wang, Y., Iqbal, M., Zheng, A., Zhao, M., Li, Z., Wang, N., Wu, C., & Yu, D. (2020). Comparative analysis of microbial community structure between healthy and Aeromonas veronii-infected Yangtze finless porpoise. Microbial cell factories, 19(1), 123. https://doi.org/10.1186/s12934-020-01383-4
- Magwaza, N.M., Nxumalo, E.N., Mamba, B.B., & Msagati, T.A.M. (2017). The Occurrence and Diversity of Waterborne Fungi in African Aquatic Systems: Their Impact on Water Quality and Human Health. International Journal of Environmental Research and Public Health, 14(5), 546. https://doi.org/10.3390/ijerph14050546
- Malinovsk, Z., Conkova, E., Vaczi, P., Harcarova, M., & Bohmova, E. (2019). Occurrence of Malassezia Spp. on Healthy Human Skin. Folia Veterinaria, 63(4), 54-59. https://doi.org/10.2478/fv-2019-0038
- Mamun, M.A.A., Sandeman, M., Rayment, P., Brook-Carter, P., Scholes, E., Kasinadhuni, N., Piedrafita, D., & Greenhill, A.R. (2019). The composition and stability of the faecal microbiota of Merino sheep. Journal of Applied Microbiology, 128(1), 280-291. https://doi.org/10.1111/jam.14468
- McPartland, J.M. (2018). Cannabis pathogens X: Phoma, Ascochyta and Didymella species. Mycologia. 86(6), 870-878. https://doi.org/10.1080/00275514.1994.12026492
- Ramette A. (2007). Multivariate analyses in microbial ecology. FEMS microbiology ecology, 62(2), 142-160. https://doi.org/10.1111/j.1574-6941.2007.00375.x
- Rawls, J.F., Mahowald, M.A., Ley, R.E., & Gordon, J.I. (2006). Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell, 127(2), 423-433. https://doi.org/10.1016/j.cell.2006.08.043
- Reema, N., Pallabi, S., Biswajyoti, B., & Pratap, P. (2018). Cutaneotrichosporon (Trichosporon) debeurmannianum: A Rare Yeast Isolated from Blood and Urine Samples. Mycopathologia, 183, 585-590. https://doi.org/10.1007/s11046-017-0231-8
- Richardson, M.J. (2008). Records of coprophilous fungi from the Lesser Antilles and Puerto Rico. Caribbean Journal of Science, 44(2), 206-214. https://doi.org/10.18475/cjos.v44i2.a8
- Sampson, T.R., & Mazmanian, S.K. (2015). Control of Brain Development, Function, and Behavior by the Microbiome. Cell Host & Microbe, 17(5), 565-576. https://doi.org/10.1016/j.chom.2015.04.011
- Shi, Y., Miao, Z.Y., Su, J.P., & Wasser, S.K. (2021). Shift of Maternal Gut Microbiota of Tibetan Antelope (Pantholops hodgsonii) During the Periparturition Period. Current microbiology, 78(2), 727-738. https://doi.org/10.1007/s00284-020-02339-y
- Shiv, M.S., Paras, N.S., Sanjay, K.S., & Prabhat, K.S. (2013). Pigment, fatty acid and extracellular enzyme analysis of a fungal strain Thelebolus microsporus from Larsemann Hills, Antarctica. Polar Record, 50(1), 31-36. https://doi.org/10.1017/S0032247412000563
- Soman, A.G., Gloer, J.B., Koster, B., & Malloch, D. (1999). Sporovexins A-C and a new preussomerin analog: antibacterial and antifungal metabolites from the coprophilous fungus Sporormiellavexans. Journal of natural products, 62(4), 659-661. https://doi.org/10.1021/np980563c
- Tian, G., Wu, X.Y., Chen, D.W., Yu, B., & He, J. (2017). Adaptation of gut microbiome to different dietary nonstarch polysaccharide fractions in a porcine model. Molecular Nutrition & Food Research, 61(10), 1700012. https://doi.org/10.1002/mnfr.201700012
- Wanasinghe, D.N., Phukhamsakda, C., Hyde, K.D., Jeewon, R., Lee, H.B., Jones, E.G., Tibpromma, S., Tennakoon, D.S., Dissanayake, A.J., Jayasiri, S.C., Gafforov, Y., Camporesi, E., Bulgakov, T.S., Ekanayake, A.H., Perera, R.H., Samarakoon, M.C., Goonasekara, I.D., Mapook, A., Li, W.J., ... Karunarathna, S.C. (2018). Fungal diversity notes 709-839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Diversity, 89, 1-236. https://doi.org/10.1007/s13225-018-0395-7
- Weber, H.A., & Gloer, J.B. (1991). The preussomerins: novel antifungal metabolites from the coprophilous fungus Preussia isomera Cain. The Journal of Organic Chemistry, 56(14), 4355-4360. https://doi.org/10.1021/jo00014a007
- Wei, S., Morrison, M., & Yu, Z. (2013). Bacterial census of poultry intestinal microbiome. Poultry Science, 92, 671-683. https://doi.org/10.3382/ps.2012-02822
- White, J.R., Nagarajan, N., & Pop, M. (2009). Statistical Methods for Detecting Differentially Abundant Features in Clinical Metagenomic Samples. PLoS Computational Biology, 5(4), e1000352. https://doi.org/10.1371/journal.pcbi.1000352
- Xu Y., Wang C., Liu H., Zhu, G., Fu, P., Wang, L., & Zhu, W. (2018). Meroterpenoids and Isocoumarinoids from a Myrothecium Fungus Associated with Apocynum Venetum. Marine Drugs,16(10), 363. https://doi.org/10.3390/md16100363
- Yang, L., Bian, G.R., Su, Y., & Zhu W.Y. (2014). Comparison of faecal microbial community of Lantang, Bama, Erhualian, Meishan, Xiaomeishan, Duroc, Landrace, and Yorkshire sows. Asian Australasian Journal of Animal Sciences, 27(6), 898-906. https://doi.org/10.5713/ajas.2013.13621
- Yang, L., Liu, S., Ding, J., Dai, R., He, C., Xu, K., Honaker, C.F., Zhang, Y., Siegel, P., & Meng, H. (2017). Gut Microbiota Co-microevolution with Selection for Host Humoral Immunity. Frontiers in Microbiology, 8, 1243-1253. https://doi.org/10.3389/fmicb.2017.01243
- Zhang, Z.Y., Chen, W.H., Zou, X., Han, Y.F., Huang, J.Z., Liang, Z.Q., & Deshmukh, S.K. (2019). Phylogeny and taxonomy of two new Plectosphaerella (Plectosphaerellaceae, Glomerellales) species from China. MycoKeys, 57, 47-60. https://doi.org/10.3897/mycokeys.57.36628
- Zhang Z.Y., Shao, Q.Y., Li, X., Chen, W.H., Liang, J.D., Han, Y.F., Huang, J.Z., & Liang, Z.Q. (2021). Culturable Fungi from Urban Soils in China I: Description of 10 New Taxa. Microbiology Spectrum, 9(2), e0086721. https://doi.org/10.1128/Spectrum.00867-21







