2021 年 3 巻 3 号 p. 72-76
2021 年 3 巻 3 号 p. 72-76
The fusion of viral and cellular membranes is an important step in infections caused by enveloped viruses. Safe and rapid cell-based assay systems have been developed to identify inhibitors of various infections caused by enveloped viruses. In this review, we have described a cell-based membrane fusion assay and its application in drug screening. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and Middle East respiratory syndrome coronavirus (MERS-CoV) induce membrane fusion by cleavage of the N-terminal region of the fusion peptide in the Spike protein by cellular proteases. To quantify the membrane fusion mediated by SARS-CoV-2 and MERS-CoV, effector cells expressing the Spike protein and target cells expressing the receptor and host protease TMRPSS2 were used. Cell fusion caused by co-culture was measured by monitoring the signals from dual split proteins (DSPs) as the reporter. Using this assay system, nafamostat was identified as a potential inhibitor of the membrane fusion caused by MERS-CoV and SARS-CoV-2. Clinical trials of nafamostat are underway. Cell-based membrane fusion assays using DSPs are expected to be useful for the discovery of viral infection inhibitors and for the evaluation of neutralizing antibodies.
• A membrane fusion assay for enveloped viruses was established using cell fusion as an indicator.
• The assay enables quantitative analysis within a few hours without using infectious viruses and is useful for the discovery of inhibitors.
• The assay revealed the inhibition activity of nafamostat, which is being evaluated as a therapeutic agent for SARS-CoV-2.
Enveloped viruses have their genetic material encapsulated by a lipid bilayer. Thus, membrane fusion between viruses and target cells is required for infection [1]. Proteins on the surface of viruses play an important role in the induction of membrane fusion. Binding of a viral fusion protein to a specific protein on the target cell membrane that acts as a receptor causes a conformational change in the fusion protein. This change activates the fusion protein, which contacts the cell membrane through a fusion peptide segment comprising hydrophobic amino acids, inducing membrane fusion.
Various fusion proteins of enveloped viruses and their receptors have been identified. In some cases, activation of fusion proteins requires cleavage by host proteases after binding to their receptors [2]. In other cases, fusion proteins are taken up by endocytosis into the cells, where structural changes due to pH changes are also important for activation [3] (Table 1). The host receptor-mediated activation of fusion proteins is a phenomenon unique to infections caused by enveloped viruses. This uniqueness provides a potent target for inhibitors of viral infections [4]. Many neutralizing antibodies inhibit viral membrane fusion by binding to fusion proteins [5]. Other compounds can also be inhibitory. For example, maraviroc (which is used to treat human immunodeficiency virus [HIV] infections) antagonizes the coreceptor CCR5 and inhibits membrane fusion mediated by an HIV fusion protein [6].
| Enveloped virus | Fusion protein | Receptor | Activator |
|---|---|---|---|
| HIV | Env (gp120, gp41) | CD4, CXCR4/CCR5 | - |
| DENV | E | LRP1 | Acidic pH |
| ZIKV | E | DC-SIGN, AXL | Acidic pH |
| SARS-CoV | Spike | ACE2 | Partial cleavage |
| MERS-CoV | Spike | CD26 | Partial cleavage |
| SARS-CoV-2 | Spike | ACE2 | Partial cleavage |
| VSV | G | LDLR family | Acidic pH |
| HCV | E1, E2 | LDLR, HSPG, SRB1, CD81, EGFR, EphA2 | Acidic pH |
| HBV | L | NTCP | Partial cleavage, Acidic pH |
| Influenza A | Haemagglutinin | Sialic acid | Acidic pH |
Enveloped viruses induce membrane fusion by binding the fusion protein to the receptor. Flaviviruses such as DENV and ZIKV require activation of their fusion proteins by acidic pH. On the other hand, coronaviruses such as MERS-CoV and SARS-CoV-2 require activation of their fusion proteins by partial cleavage after binding to the receptor. In addition to the expression pattern of the receptors, the activation mechanism of the fusion protein is important for target organ selection. HIV, human immunodeficiency virus; DENV, Dengue virus; ZIKV, Zika virus; SARS-CoV, severe acute respiratory syndrome coronavirus; MERS-CoV, Middle East respiratory syndrome coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; VSV, vesicular stomatitis virus; HCV, hepatitis C virus; HBV, hepatitis B virus.
We established a cell-based fusion assay for fusion proteins of various enveloped viruses, including HIV [7, 8], flaviviruses that include dengue virus and Zika virus (ZIKV) [9], and coronaviruses that include Middle Eastern Respiratory Syndrome (MERS)-CoV [10] and Severe Acute Respiratory Syndrome (SARS)-CoV-2 [11], to evaluate the antiviral activities of chemical compounds and antibodies against enveloped virus infections.
The cell-based membrane fusion assay used effector cells and target cells. Effector cells express a fusion protein derived from an enveloped virus. In contrast, target cells express their host receptors. Upon co-culture, the interaction between the fusion protein and the receptor results in the formation of a fused cell syncytium (Fig. 1A). To quantify syncytium formation, dual split proteins (DSPs) were used as reporters. DSP1-7 and DSP8-11 were developed by Dr. Zene Matsuda and colleagues [12] (Fig. 1B). The DSPs are composed of fragments derived from Renilla luciferase (RL) and green fluorescent protein (GFP). In isolation, the DSPs do not show any enzymatic or fluorescence activity. However, the GFP portions of the DSPs self-associate and induce the association of the RL portions when both DSPs are expressed in the same cell. This reassociation of the DSPs restores the RL enzymatic and GFP fluorescence activities. Therefore, cell-cell fusion between effector cells with one DSP and target cells with the other DSPs can be easily monitored by detecting RL and/or GFP signals. GFP fluorescence signals allow qualitative image analysis of membrane fusion by fluorescence microscopy, while RL signals are useful for more quantitative measurements of membrane fusion by monitoring the RL activities with a luminometer. This cell-based membrane fusion assay has several advantages over conventional methods that use intact infectious viruses and pseudo-type viruses. First, the cell-based membrane fusion assay does not require high-level biosafety facilities, such as Biosafety Level (BSL) 2, BSL3 or BSL4. For example, the analysis of SARS-CoV-2 or MERS-CoV requires BSL3 facilities, equipment, and skilled operators. Recently, in order to avoid these complicated safety measures, chimeric vesicular stomatitis virus (VSV) [13] with fusion genes and pseudoviruses with fusion proteins on their envelopes similar to HIV [14] and VSV [15, 16] have been widely used for analysis. Pseudoviruses lack some or all of the structural genes and cannot replicate, which allows for the relatively safe analysis of the entry stage of the virus. However, because these recombinant viruses are infectious to humans, BSL2 or BSL3 facilities are required to handle them. Moreover, advanced techniques are required to prepare pseudoviruses with sufficient infection efficiency. Alternatively, the cell-based fusion assay uses HEK293T cells, which have stable proliferative capacity and can be cultured using a simple method. Once a stable cell line expressing the fusion protein is established, the effector cells can be easily prepared for the assay by treatment with appropriate antibiotics, and no specific equipment or techniques are required. Second, the RL and GFP signals can be obtained in a short period of time because fused cells are formed within a few hours, leading to the rapid observation of cell-cell fusion. Third, RL and GFP signals can be analyzed in living cells using a membrane-permeable substrate for RL without additional steps, such as preparation of cell lysates or fixation and staining of cells. These factors make it easy to apply the assay to the screening of many samples, such as high-throughput screening. In addition, in classical viral infection experiments using reporter gene expression systems, toxic compounds that have non-specific effects on nucleic acid synthesis or protein translation are often obtained as false positives, for example, due to the reduction of reporter gene expression levels. On the other hand, in our membrane fusion assay, fusion proteins, receptors, and split reporters were fully expressed in each cell when the cells were co-cultured, thus reducing false positives caused by non-specific inhibitors of molecular synthesis. As described above, the cell fusion assay has various advantages, but there are some points that should be noted while establishing cells for this assay. The expression level of the fusion protein is critical for a successful assay, especially for the establishment of effector cells. Codon optimization of the fusion protein and promoter selection are particularly important. Viral antigens are generally regulated to maintain low expression, and native codon sequences may not induce expression at levels sufficient for cell fusion. In addition, the fusion genes may be affected by splicing in the nucleus. Codon optimization may disrupt RNA structure and adversely affect its function, but it may help if the cell fusion signal is low. It is also preferable to select a strong promoter such as the CMV or EF1α promoter. However, because the overexpression of membrane proteins can induce cellular stress, the optimal promoter depends on the specific fusion protein. For example, the expression of the HIV env gene by the retroviral vector pMXs [17] is sufficiently high for cell fusion, while the SARS-CoV-2 Spike gene is expressed efficiently by the EF1α promoter, which allows for efficient cell fusion. In the case of the SARS-CoV-2 Spike gene, induction by the CMV promoter resulted in relatively low expression efficiency with a lower cell fusion signal compared with that induced by the EF1α promoter.
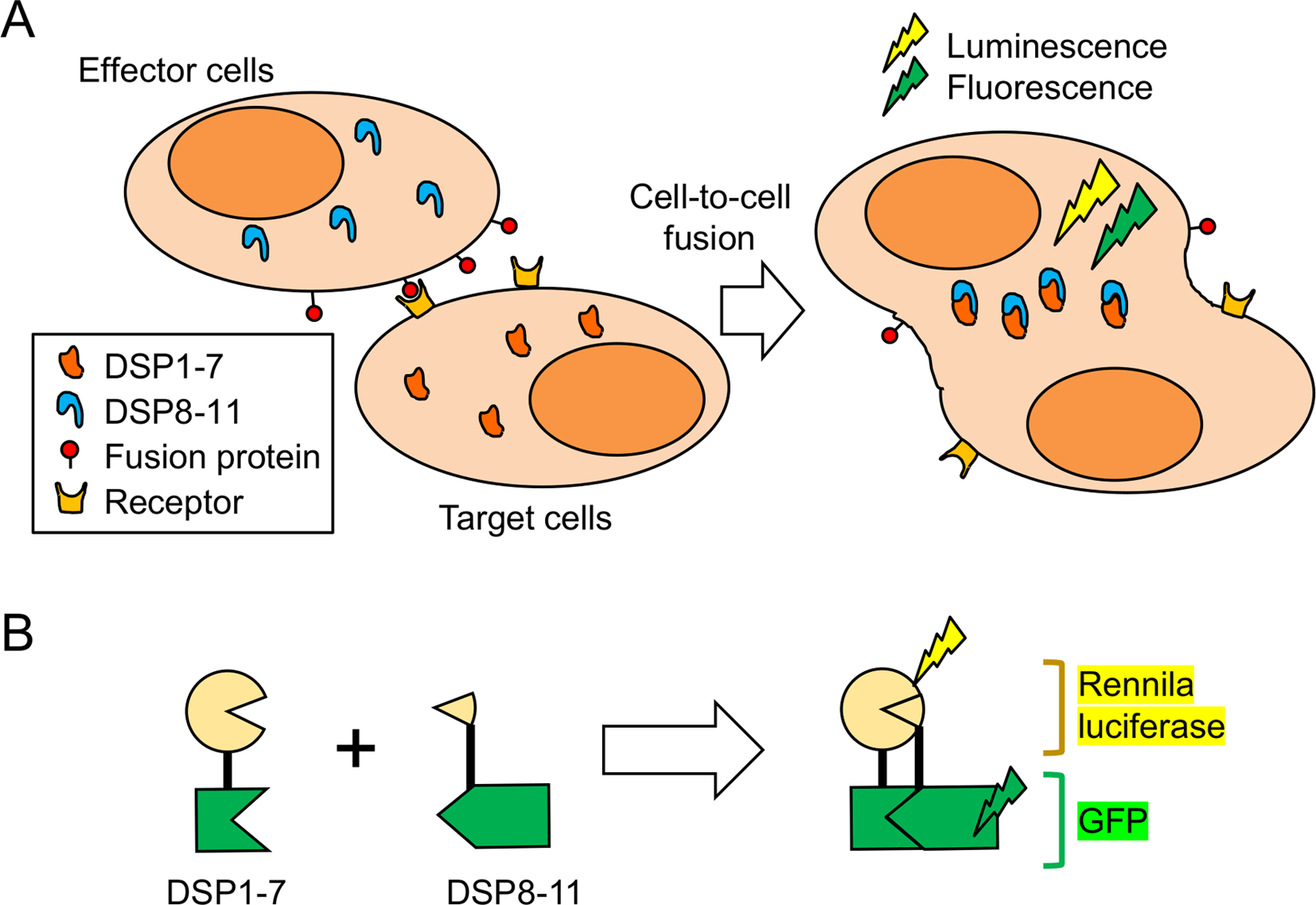
Call-based assay for fusion protein-mediated membrane fusion using dual split proteins (DSPs). A. Scheme of the membrane fusion assay. Fusion cells formed by co-culturing effector cells expressing the fusion protein and target cells expressing the receptor are quantified using the association of a split reporter protein. B. Structure of the split reporter protein DSPs. DSP1–7 has the structure Renilla luciferase [1–155]-Ser-Gly-Gly-Gly-Gly-GFP [1–156]. DSP8–11 has the structure Met-GFP [157–231]-Gly-Gly-Gly-Gly-Ser-Renilla luciferase [156–311]. Renilla luciferase and GFP become active only when DSP1–7 associates with DSP8–11.
In 2015, with the support of the Japan Agency for Medical Research and Development (AMED), we developed an assay system that can analyze the membrane fusion of MERS-CoV. This coronavirus has a 40% mortality rate [18]. An outbreak of MERS occurred in South Korea in 2015. The Spike fusion protein of MERS-CoV recognizes the CD26 receptor, which allows the infection of airway epithelial cells. Importantly, after binding to the receptor, the N-terminal side of the fusion peptide is cleaved by two major host proteases to induce membrane fusion. One is the serine protease TMPRSS2, which is expressed in the plasma membrane. The other is the endosomal cathepsin L, which is expressed in the endosome. In airway epithelial cells, TMPRSS2, which acts on the cell membrane, has been suggested to be important for infection. Subsequent analysis of TMPRSS2-knockout mice showed that cleavage of Spike by TMPRSS2 is important for MERS-CoV pneumonia symptoms [19]. We established cells expressing the CD26 and TMPRSS2 as target cells and co-cultured them with effector cells expressing the MERS-CoV Spike protein [10]. The membrane fusion could be quantified by measuring RL signals 4 hr after co-culture. To screen existing drugs for MERS-CoV infection inhibitors, we applied this membrane fusion assay to screen Food and Drug Administration (FDA)-approved drug libraries. The screen revealed that the serine protease inhibitor nafamostat suppressed membrane fusion by inhibiting TMPRSS2 protease activity. Nafamostat is a potent inhibitor of coronaviruses at only 10% of the concentration of camostat, a TMPRSS2 inhibitor that has already been reported to inhibit coronavirus infection. Infection experiments with MERS-CoV demonstrated that nafamostat inhibited infection of airway epithelial cell-derived Calu-3 cells at approximately 1 nM. Since it is known that the blood concentration of the drug can be maintained at 30–200 nM by continuous infusion [20], it is conceivable that the drug could inhibit coronavirus infection.
COVID-19 is caused by infection with SARS-CoV-2 [21]. COVID-19 quickly spread around the world soon after the first patient was identified in Wuhan, China, at the end of 2019. By April 2021, approximately 140 million people worldwide were infected with SARS-CoV-2, and more than 3 million people had died [22]. The pandemic is ongoing.
We established a membrane fusion assay using the SARS-CoV-2 Spike protein and its receptors, ACE2 and TMPRSS2, to investigate the effect of nafamostat on membrane fusion mediated by the Spike protein. Nafamostat strongly suppressed SARS-CoV-2 infection as well as MERS-CoV at concentrations lower than the concentrations that can be achieved in circulating blood. Since SARS-CoV-2 infection also causes systemic organ damage due to thrombosis, nafamostat may have potential therapeutic value via inhibition of viral entry and thrombosis.
Clinical trials of the combination therapy with nafamostat and the viral RNA polymerase inhibitor favipiravir for COVID-19 are currently being conducted in Japan [23, 24]. The development of a new nafamostat formulation that acts directly on airway epithelial cells by changing the route of administration from conventional injection to inhalation is also underway [25].
In addition to inhibitors of membrane fusion, drugs that inhibit viral replication and the production of functional viral proteins in infected cells are being developed [26, 27]. The combination of nafamostat and other membrane fusion inhibitors with inhibitors that target different steps of viral infection is expected to lead to therapies with fewer side effects and less emergence of viral resistance. We hope that our membrane fusion assay will be useful for rapid drug development.
The authors declare no conflicts of interest associated with this manuscript.
This work was supported in part by grants-in-aid from the Japan Society for the Promotion of Science (18K15235 and 20K07610 to MY), and by the Japan Agency for Medical Research and Development (AMED) [Program of Japan Initiative for Global Research Network on Infectious Diseases (JGRID) JP18fm0108006 and JP20wm0125002 to JG].