2022 年 19 巻 論文ID: e190005
2022 年 19 巻 論文ID: e190005
How can evolution assemble lifeless molecules into a complex living organism? The emergent process of biological complexity in the origin of life is a big mystery in biology. In vitro evolution of artificial molecular replication systems offers unique experimental opportunities to probe possible pathways of a simple molecular system approaching a complex life-like system. This review focuses on experimental efforts to examine evolvability of molecules in vitro from the pioneering Spiegelman’s experiment to our latest research on an artificial RNA self-replication system. Genetic translation and compartmentalization are shown to enable sustainable replication and evolution. Latest studies are revealing that coevolution of self-replicating “host replicators” and freeloading “parasitic replicators” is crucial to extend evolvability of a molecular replication system for continuous evolution and emergence of diversity. Intense competition between hosts and parasites would have existed even before the origin of life and contributed to generating complex molecular ecosystems. This review article is an extended version of the Japanese article “An in vitro evolutionary journey of an artificial RNA replication system towards biological complexity” published in SEIBUTSU-BUTSURI Vol.61, p.240–244 (2021).”
No one has witnessed evolution of non-living matter into complex living organisms. In vitro evolution enables direct observation of how a molecular replication system builds up complexity through evolution. This review describes a brief history of in vitro molecular evolution and essential factors for emergence of complexity. Above all, our recent findings indicate that coevolution with molecular parasites is a key to allow a molecular replication system to undergo continuous evolution and diversification toward higher complexity.
Can you believe that life has spontaneously arisen from a mixture of non-living molecules? Origin of life studies and evolutionary biology are revealing that all organisms date back to the last universal common ancestor (LUCA) [1–3], and a molecular world centered on RNA-like polymers further predated the origin of life [4–8]. This abiogenesis view, however, apparently conflicts with the conclusion of the famous Pasteur’s swan flask experiment that disproved the spontaneous generation of life in the absence of parent organisms. How can we understand the gap between these contradictory views? Is it true that organic molecules self-organized into a living cell in the ancient earth? There are no fossil records of prebiotic molecular replication systems, and genomics and molecular evolution approaches cannot access the history before LUCA. Do we have any means to investigate this hypothetical origination of life from non-living molecules, which has never been witnessed?
A famous quote “What I cannot create, I do not understand” by Richard Feynman would be true of living systems. In fact, we are in the unprecedented international boom of artificial cell synthesis campaigns [9–12], where different fields of scientists get together to test their understanding of the biological system by bottom-up construction of the cell. Though there is great progress, we still have a long way from the clear understanding and rational design of even the simplest form of life as bacteria.
The key concept in this review is “evolutionary design” instead of rational design of a full-fledged cell. Extant living systems would not have appeared in a moment by a single historical incident on earth. It is the consequence of billions of years of evolution, which must have polished up much simpler molecular replication systems into present organisms. Therefore, we propose to understand how complexity originates through in vitro evolution of a minimal replication system and analysis of its evolutionary processes. If life has really evolved from a mixture of molecules, we should be able to reproduce the essence of the complexification process of the molecular system developing toward life. Troubleshooting during this experimental evolution and modification to the design of the replication system will pose us a new point of view and we will better understand how difficult hurdles are to be overcome. In this review, we present a brief history of in vitro molecular evolution approaches focused on artificial RNA replication systems derived from bacteriophage Qβ. We start from the pioneering Spiegelman’s experiment and go on to describe our recent studies, discussing what is required for a molecular replication system to approach biological complexity.
Evolution is a process in which heritable properties of a population change over generations. Evolution requires three processes:
1. Replication: individuals make their copies and offspring inherit the genetic information (genotype)
2. Mutation: heritable genotypes diversify by stochastic errors in replication or environmental perturbation
3. Selection: chances of survival are biased depending on observable properties (phenotype) that are determined by genotypes
In the biological world, replication and selection cannot be decoupled and happen simultaneously in general. For example, when organisms are under continuous pressure to eliminate them (e.g. predators to eat them), selection occurs upon reproduction events due to variable number of offsprings and survivors to change the genetic composition of a population. This type of evolution is often referred to as “Darwinian” evolution with natural selection. Note that, however, evolution is not a concept limited to biological organisms, but any population satisfying the three requirements is able to evolve. Up until the mid 20th century, however, no one had witnessed evolution of non-life.
In 1967, the first in vitro Darwinian evolution was reported by Spiegelman’s group [13]. They extracted the genomic RNA from a single-stranded RNA virus (bacteriophage Qβ) and added it to a reaction mixture containing nucleotides and the RNA polymerase of Qβ (Qβ replicase) to replicate the genomic RNA as templates (Figure 1). Evolution was expected to occur in the serial transfer because the RNA population should diversify through random mutations introduced in the successive replication, and fast replicating RNA mutants propagate and gradually replace the original RNA population. They successfully demonstrated the proof-of-concept: after 74 rounds of passages, RNAs that replicate 15 times faster than the original RNA evolved. This cornerstone study has shown for the first time that molecules have evolvability (ability to evolve) out of living organisms.
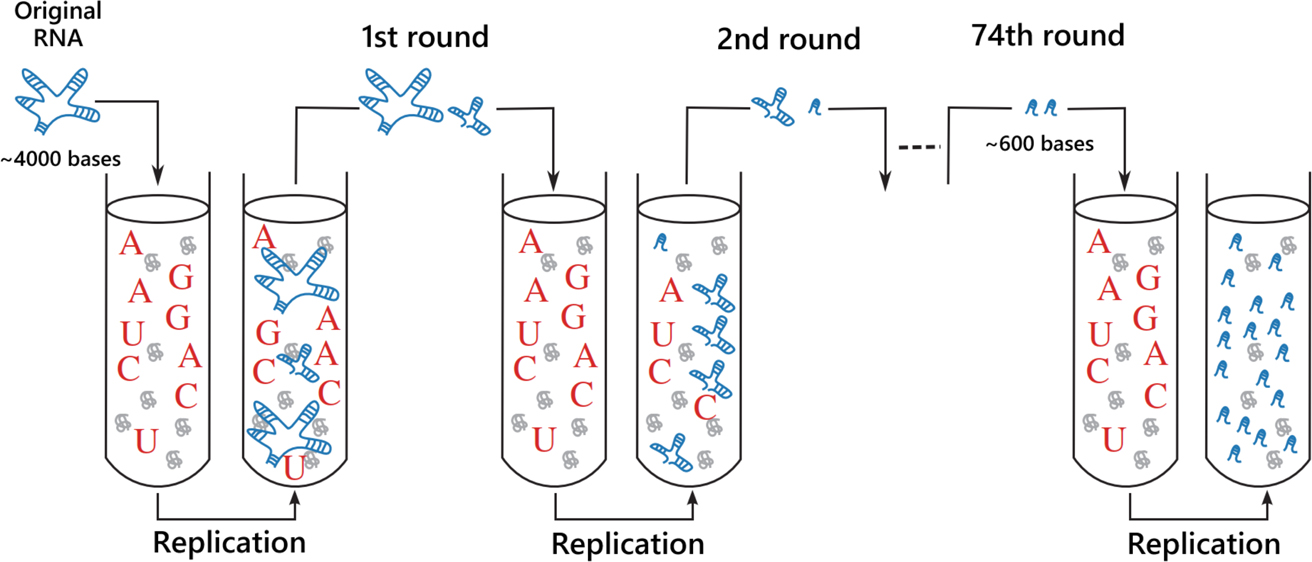
Schematic of Spiegelman’s experiment. Qβ replicases (grey globules), nucleotides (red alphabets) were prepared in test tubes. The genomic RNA (blue ladders) was inoculated in the beginning, then serially transferred. RNA molecules evolved faster and shorter during the experiment. (the figure modified from Takeuchi et al. [14])
Does this mean that a further long-term experiment will evolve the RNA replicators into a complex system like living organisms? The answer is probably no. In fact, the original RNA (~4000 bases) became shorter and shorter during the serial transfer and finally simplified to only 600 bases long (so-called Spiegelman’s monster). This was probably because the only selection pressure for RNA in this experiment was to become more efficient replication templates, and the original genomic information for viral infection was only a burden for replication and easily discarded by mutations. In addition, the evolution of the RNA stagnated after 74 rounds of passages, converging to the sequence with 600 bases.
After Spiegelman’s experiment, another group reported evolution of a DNA-based replication system [15], and the length of DNA templates gradually shortened and stopped changing as in Spiegelman’s experiment. These results may indicate the low evolvability of these replication systems. In a nutshell, replication of the templates (RNA or DNA) tends to be faster as sequences become shorter, so once the sequence gets short enough to function as a template, it is optimal and does not evolve anymore. If that is the case, what is required for further evolution?
A hallmark of life is self-replication instructed by its genetic information. This seems to be a distinguishing property of a living system missing in Spiegelman’s RNA replication system. In Spiegelman’s experiment, the replicase (i.e. replication function) was provided in the reaction mixture by experimentalists instead of being produced by translation of the self-encoded genetic information in RNA. On the other hand, living organisms can “self-replicate” by the translation of replication functions encoded in their genomes. What if we introduce the self-replication function in Spiegelman’s system via translation of the replicase gene? Reductive evolution of RNA may be prevented by this design because RNA needs to provide the replicase by themselves from the intact replicase gene. In this case, RNA can evolve the self-encoded replicase information in addition to the template information. Therefore, the introduction of self-replication ability is expected to make a significant difference in evolvability of the replication system. Please note that the essence here is the introduction of self-replication based on its genetic information, and translation is just one way to implement this function.
Based on this idea, we constructed a translation-coupled RNA replication system [16] (Figure 2) by introducing a cell-free translation system (a solution containing translation proteins, tRNAs, NTPs, and amino acids) [17] to the Spiegelman’s RNA replication system. Note that the cell-free translation system does not contain the replicase, and RNA replicators need to provide the replicase via translation of its gene. To test evolvability of the new RNA replication system with translation ability, we performed a serial transfer experiment.
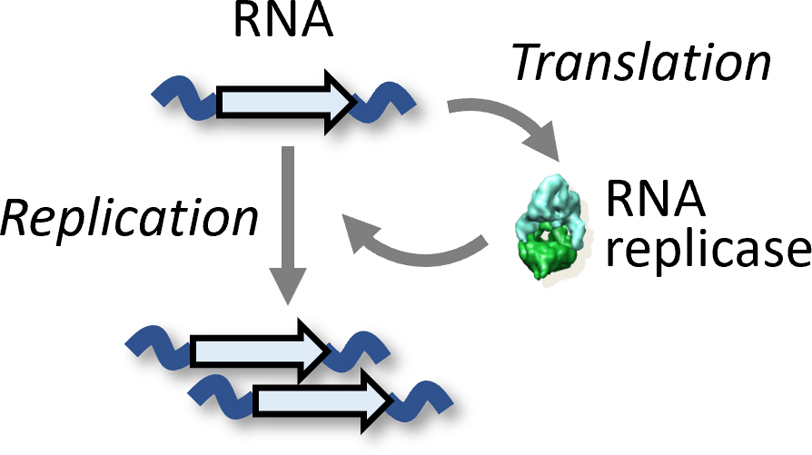
Schematic of the translation-coupled RNA self-replication system. The RNA replicators self-encode the replicase and replicate via cell-free translation of the replicase.
Unfortunately, the first evolution experiment of the translation-coupled RNA replication system did not work. During the serial transfer experiment, the average concentration of RNA replicators gradually decreased and went under the detection limit after 17 rounds. Why did it fail? We noticed there were two entangled reasons. First, mutations are mostly deleterious and more common than beneficial ones to improve gene function [18]. Therefore, the RNA population was gradually dominated by inactive RNA mutants that lost genetic information of the active replicase. But here you might get confused: why better RNA mutants do not propagate, regardless of improved function for faster replication?
The second reason was the emergence of parasites that prevented the propagation of improved host RNA mutants. The point is that RNA replicators easily lose the active replicase information by random mutations, but many of them can still work as templates for replication. Therefore, if RNA replicators exist in the same bulk solution, template RNAs with broken genes (parasitic RNAs) can freeride replicases translated by RNAs encoding the active replicase genes (host RNA). This situation is a nightmare for the host RNA because not only the parasitic RNA freeloads, but also can discard unnecessary genetic information to quickly become smaller and efficient templates that badly inhibit the replication of the host RNA [19]. In fact, we experimentally confirmed that small RNAs, similar sequences to Spiegelman’s monster, actually appeared and dominated the RNA population. Then how can we protect the host RNA to enable sustainable replication for evolution?
The parasite problem has been a focus in the origin of life studies from decades ago, and compartmentalization by a cell-like structure was theoretically suggested as a solution to mitigate the threat of parasite invasion [20–23]. What difference does compartmentalization make in the case of the translation-coupled RNA replication? If we segregate all RNA replicators in different compartments (i.e. one copy per compartment), an active host RNA mutant translates the replicase (phenotype) from its RNA sequence (genotype) and performs clonal amplification of itself without parasitic replication (Figure 3). On the other hand, inactive RNA mutants cannot replicate because there is no way to access active replicases. Therefore, this genotype-phenotype coupling by compartmentalization offers a positive selection mechanism for active host RNA mutants. Moreover, even when fast replicating small parasites appear somewhere, they cannot propagate to other compartments due to physical segregation. For these reasons, we attempted to introduce compartmentalization to our translation-coupled RNA replication system.
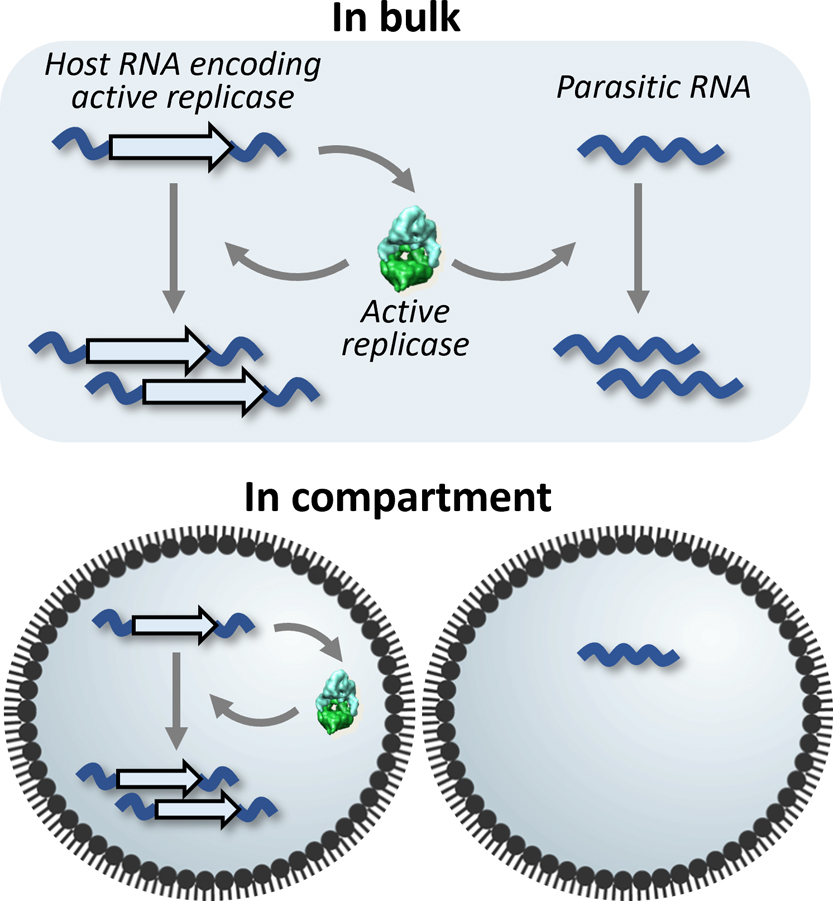
Effect of compartmentalization. Under the non-compartmentalized bulk condition (top), a parasitic RNA can replicate by using the replicase translated from a host RNA, whereas under the compartmentalized condition (bottom), if each RNA is isolated into different compartments, a parasitic RNA cannot replicate because it cannot utilize the replicase.
We achieved compartmentalization using a water-in-oil emulsion method [24]. We dispersed a cell-free translation system containing only the host RNA in mineral oil with surfactants, then subjected them to a serial transfer experiment (Figure 4A). We conducted limiting dilution of RNA replicators to guarantee one copy encapsulation of RNAs before replication reaction when the emulsion was transferred every round. As expected, the host RNA population continued to replicate and evolved self-replication ability more than 100-fold without losing the replicase gene (Figure 4B) [25]. Notably, we found that both template (RNA sequence) and replicase (protein sequence) information coevolved to achieve the high self-replication ability. In conclusion, the introduction of genetic translation and compartmentalization enabled evolution with higher complexity than in the Spiegelman’s experiment.

Serial dilution experiment without parasitic RNAs. (A) Schematic of the serial dilution experiment. The host RNA and the reconstituted translation system are encapsulated into water-in-oil droplets and incubated at 37°C for 1–2 h for translation of the RNA replicase and RNA replication by the replicase. An aliquot of the droplets is then diluted with new droplets containing the translation system and vigorously mixed to induce fusion and division of the droplets. The resultant droplets are incubated again for the next round of translation and replication. (B) Trajectory of the replication efficiency of the RNA population. The replication efficiency refers to the RNA concentration after replication with initial 0.1 nM RNA. The efficiency dropped at the points indicated with the arrowheads, at which experimental methods are changed slightly (i.e., RNA recovery method was changed at the round 32 and the incubation time was shortened at the round 80). (C) Trajectory of the average number of mutations accumulated in the RNA. Figures are modified form Ichihashi et al 2013 [25].
Now can we expect that the RNA replication system is going to evolve into a more complex system? The answer is probably no again. The evolution of this self-replicating system gradually slowed down, and after about 600 generations, both the improvement of replication rate and the accumulation of mutations almost stopped (Figure 4B and 4C). The RNA population converged to a single host RNA with little diversity [26], and the length of the RNA sequence did not significantly change. It appeared that this RNA replication system reached evolutionary stasis too, indicating the current setup was still not sufficient to produce rich evolutionary dynamics. What is still missing to achieve open-ended evolution and diversification as living organisms do?
Parasitic entities as viruses are well-known to be important players in biological evolution [27–29]. Especially, the evolutionary arms race is a key concept [30–33], in which hosts and parasites compete to avoid infection and parasitize efficiently, thus promoting mutual adaptive evolution. Indeed we can see how parasites are influential to evolution by the fact that even simple organisms as bacteria and archaea have complex defense mechanisms, such as the CRISPR-Cas system [34].
Let’s look back at the setup in the previous evolution experiment in the light of parasites. We simply considered parasitic RNA as inhibitory factors and used compartmentalization to cut the interaction between host and parasitic RNAs. Now, what will happen if we can do evolution with strong interaction of the two?
Under compartmentalization, it is possible to conduct an evolution experiment in which host and parasitic RNAs coexist. In the previous serial transfer experiment, the number of RNAs was controlled to be smaller than the number of compartments. We can remove this limitation and just do serial transfers under the condition where multiple copies of host and parasitic RNAs coexist in a compartment. In this case, we will have a rapid increase of parasites in the beginning and they will inhibit the replication of the host RNA, resulting in a decrease in host RNA concentration. If we continue serial transfers, however, the concentration of parasitic RNA will also decrease following the host RNA because parasites cannot replicate without their hosts. When the total number of RNA replicators becomes less than the number of compartments, single-molecule encapsulation is achieved again, and the host RNA will be able to amplify again without parasitic amplification. As the number of hosts increases, the number of parasites also increases again, and RNA replication is expected to continue with oscillatory population dynamics of both host and parasitic RNAs (the authors investigated possible behaviors using a mathematical model [35]). Expecting coevolution of host-parasite brings something new, we performed a serial transfer experiment again under the condition of host-parasite coexistence.
The initial host RNA in this coevolution experiment was a sequence taken from the previous experiment that almost stopped evolution [25], but mutation accumulation restarted and the host RNA kept evolving throughout the serial transfer. In the early stage of this experiment, we observed oscillation dynamics in the host and parasite RNA concentrations with a period of about 10 rounds (Figure 5A). The amino acid composition of the replicase encoded in the host RNA changed around 40 round, and the evolved replicase preferentially replicated the host RNA over the original parasitic RNA (parasite-α) [36]. The oscillation pattern became unclear after this, and new parasitic RNAs (parasites-β and -γ) with significantly different sequence lengths appeared after 90 round [37] (Figure 5A). Surprisingly, these parasites had longer sequences than parasite-α that appeared early in evolution (Figure 5B). This was an interesting observation against the previous evolution in which parasites only evolved to become shorter.
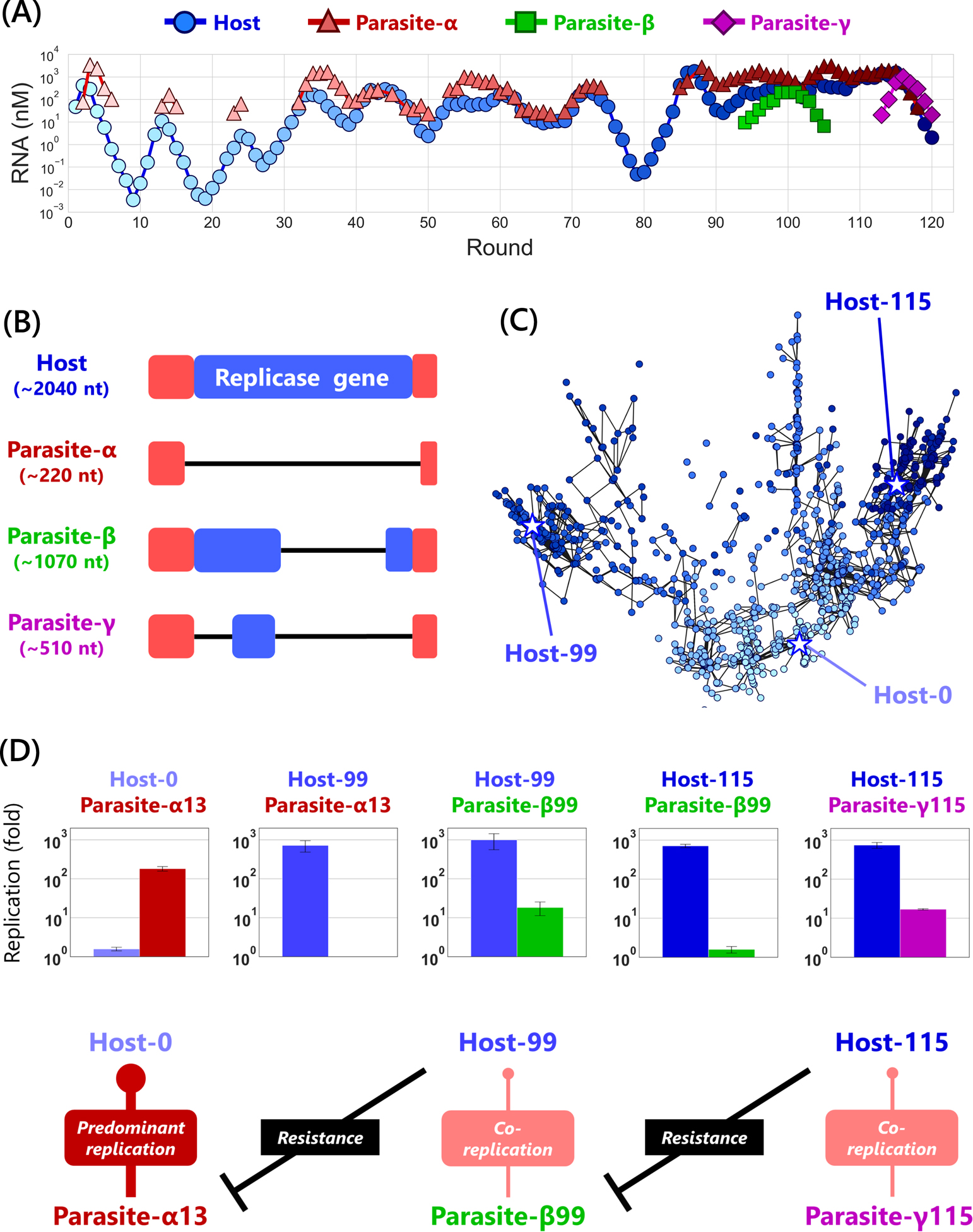
Evolutionary dynamics of the host and parasitic RNAs. (A) Population dynamics of the host and parasitic RNAs during the serial transfer. (B) Schematic of the alignments of the host and parasitic RNAs. The replicase gene and the recognition sequence of the replicase are represented in blue and red respectively. (C) 2D representation of the dominant genotypes of the host RNA. A point represents one genotype. The color depths are consistent with those in (A). Two points connected by a black line are one Hamming distance apart. Stars represent the genotypes of the evolved RNAs used for the competitive replication experiments in Figure 5D. (D) Competitive replication experiments. Host-0 is the original host RNA. Evolutionary arms races between hosts and parasites are clearly shown.
The figures are modified from Furubayashi et al. 2020 [37].
In addition, the evolutionary pattern of these RNA populations was remarkably rich in diversity. Using next-generation sequencers, we performed a comprehensive analysis of RNA sequences during the evolution and revealed that the host RNAs diverged into multiple lineages in a sequence space [36] (Figure 5C). This was in sharp contrast to the previous (almost) parasite-free evolution that generated the unidirectional evolutionary pattern poor in diversity [26]. The parasitic RNAs also continued to evolve throughout the experiment. Parasite-α appeared in the early phase survived and accumulated unique mutations not found in the host RNA population. These mutations unique to parasites indicate that a parasite population might work as a sequence pool to generate and store new sequence information. The newly emerged parasites-β and -γ had identical sequences to those of host RNAs that coexisted at the time of their appearance (rounds 99 and 115), suggesting that they originated from different hosts independently.
To directly examine whether host-parasite relationships coevolved, we conducted a series of competitive replication experiments between isolated clones of evolved host and parasitic RNAs. Specifically, we synthesized the representative sequence of the original, 99-round, and 115-round host RNA and the parasite-α, -β, and -γ RNAs. We made host-parasite pairs by picking up a sequence from the host and parasitic RNAs and performed the translation-coupled replication reactions in a cell-free translation system for each pair. We found that the host RNA, which was initially outcompeted by the parasite RNA, evolved to suppress the parasitic replication, but the parasitic RNA evolved to increase replication rate again, resulting in mutual adaptive evolution in turn. That is, the evolutionary arms race in vitro was demonstrated (Figure 5D) [37].
What enabled continuous and diversifying evolution of the RNA? It was probably due to the dynamic environment introduced by evolving parasitic RNAs. In the previous evolution experiment, the environment was constant for the host RNA because it always replicated from a single copy in the same composition of the cell-free translation system. In this situation, the evolution probably stopped when the optimal sequence for that environment was discovered. In the coevolution experiment, on the other hand, coevolving parasitic RNAs offer dynamic environments for host RNAs. An optimal host RNA in a specific environment becomes not optimal due to freeride by newly evolved parasitic RNAs. Thus, coevolving host and parasitic RNAs play cat and mouse in a sequence space, and evolution would not converge easily. Since parasites inevitably appear in systems above a certain complexity [38,39], they could have played a pivotal role for continuous evolution and diversification of primitive replication systems that approached more complex cellular organisms.
In vitro molecular evolution has contributed to understanding evolvability of molecules. First, self-encoded information (gene) and a self-replication function instructed by the gene are necessary to evolve without the loss of information. This would give the starting point of a “self-replicating” molecular system, a requirement of the living system. Second, compartmentalization enables genotype-phenotype coupling and prevention of parasite propagation. Without the two functions given by compartmentalization, a replication system cannot undergo positive selection, nor sustainable replication for long-term evolution. Finally, coevolution with parasites enables continuous evolution and diversification of a replication system driven by mutually evolving factors, hosts and parasites. As described in this paper, through experimental evolution and modification of a replication system, we can identify important factors for molecules to evolve toward more complex systems.
We would like to complement our arguments above to avoid possible misunderstanding. First, the cell-free translation system was used to implement the self-replication function in the presented studies, but we used it for practical reasons and translation is not the required condition to achieve evolution of complexity. A more primitive replicase ribozyme [40,41] can replace this function in principle and is assumed to be the workhorse before the origin of life [7,8]. Second, the RNA replicator encoding Qβ replicase (~2000 bases) is just an artificial experimental model to demonstrate Darwinian evolution in vitro and far from the primitive replicators assumed to be less than 100 bases [42]. Since an evolutionary trajectory of self-replicators can hugely depend on the initial sequence, physicochemical properties of molecules, and surrounding environments, more varieties of self-replication systems and evolutionary setups are necessary to further investigate primitive molecular evolution.
Where does the RNA evolution go in the end? The evolution of the translation-coupled RNA replication system is still ongoing, and RNA replicators have become even more complex to form a multiple membered host-parasite network [43]. We expect “cooperation” among RNA replicators is the next step for the system to approach biological complexity. Cooperation among different elements is the universal structure prevalent in the biological hierarchy from molecule to ecosystem levels, essential for novel functions and structures. Are RNA replicators going to find new relationships to complement mutual survival after the fierce competition? Or will they eventually stagnate again, unable to escape from endless competition for survival? No one knows the conclusion yet. An in vitro evolutionary journey has just begun.
The author declares no conflict of interest.
T.F. and N.I wrote the manuscript.