2019 Volume 1 Issue 11 Pages 487-492
2019 Volume 1 Issue 11 Pages 487-492
Variant types and sites in a single gene could influence the age of onset, severity, and pattern of affected organs of the genetic disease, such as in Marfan syndrome (MFS)-causing FBN1, and understanding the genotype-phenotype relationship could aid in determining the treatment strategy. In contrast, completely distinct system and/or organ diseases induced by 1 gene mutation have been rarely reported. Transforming growth factor-β (TGF-β) type I receptor-encoding TGFBR1 is such a gene, causing Loeys-Dietz syndrome (LDS) closely related to MFS, and also multiple self-healing squamous epithelioma (MSSE) without clinical overlap. The detailed mechanisms underlying this effect, however, remain elusive. We recently reported the significance of 2 distinct intronic variants (c.973+1G>A and c.806-2A>C) of TGFBR1, which were both predicted to mediate in-frame exon 5 skipping but caused LDS and MSSE, respectively. On ex vivo minigene splicing assay analysis we demonstrated that 2 different cryptic splice sites were activated, and in-frame and out-of-frame transcripts were produced in LDS and MSSE, respectively, supporting the previously proposed but not yet approved mechanism that loss-of-function and haploinsufficiency-causing variants in serine/threonine kinase domains induce LDS and MSSE, respectively. In this review, we briefly summarize the recent findings and unresolved problems for the pathogenesis of LDS, including the TGF-β signaling paradox: most variants have been verified or predicted to be loss of function in vitro, but these variants enhanced TGF-β signaling in vivo.
Loeys-Dietz syndrome (LDS) is an autosomal dominant heritable disorder of the connective tissue closely related to Marfan syndrome (MFS), which is characterized by a triad of arterial tortuosity and aneurysm, widely spaced eyes (hypertelorism), and bifid uvula. Patients with LDS are more likely to exhibit rapidly progressive aortopathy with a tendency to rupture and dissection in the aorta/arteries at a young age and at smaller dimensions compared with MFS.1–3 LDS is caused by a pathogenic variant in transforming growth factor-β (TGF-β) signaling-related genes and classified according to the pathogenic genes: TGFBR1 (LDS1), TGFBR2 (LDS2), SMAD3 (LDS3), TGFB2 (LDS4), TGFB3 (LDS5), and SMAD2 (LDS6). A large proportion of LDS patients have pathogenic variants in the genes encoding TGF-β types I and II receptors, TGFBR1 (20–25%) and TGFBR2 (55–60%), especially in the serine/threonine kinase (STK)-encoding regions.4,5 Most variants have been verified or predicted to cause STK loss of function in vitro, but these variants enhanced TGF-β signaling in the in vivo aortic regions, referred to as the TGF-β signaling paradox.
In contrast, pathogenic variants in TGFBR1 also induce a distinct skin disease called multiple self-healing squamous epithelioma (MSSE).6 MSSE is an autosomal dominant skin cancer syndrome characterized by the development of multiple rapidly growing invasive skin tumors that emerge for a few weeks only to subsequently spontaneously regress and heal with scarring, and there is no clinical overlap between LDS and MSSE.
We recently reported a familial Japanese case of LDS with a novel splice donor site variant in intron 5 in TGFBR1 (c.973+1G>A).7 The in-frame deletion of the whole exon 5 was predicted to be elicited by this variant, whereas a variant of MSSE in a British family (c.806-2A>C) was also predicted to induce the same deletion of exon 5.6 To clarify this, we performed minigene-based splicing assays of both variants and found that these 2 different diseases were caused by differences in the splicing patterns.
In this mini review, we present and discuss the recent understanding of the molecular mechanism of LDS and the unresolved problems, including the TGF-β signaling paradox and the mechanism by which TGFBR1 variants elicit 2 completely distinct diseases, LDS and MSSE.7
TGFBR1 and TGFBR2 are transmembrane STK receptors consisting of 9 and 7 exons, respectively. The activation of TGFBR2 by TGF-β ligands induces TGFBR2 dimerization, and the TGFBR2 homodimer forms a stable receptor complex with TGFBR1 homodimer and phosphorylates TGFBR1, leading to the subsequent activation of the SMAD signaling pathway. Phosphorylated SMAD2 and SMAD3 form stable complexes with SMAD4, which translocate to the nucleus and regulate the transcription of target genes (Figure 1A).8 In LDS, most variants in TGFBR1/TGFBR2 are missense and are located in or immediately flanking the evolutionarily conserved STK domain.9 Recently reported knock-in mice with missense mutations (Tgfbr1M318R/+ and Tgfbr2G357 W/+) identically developed vascular, craniofacial, and skeletal manifestations of LDS, but heterozygous knockout mice (Tgfbr1+/− and Tgfbr2+/−) did not develop any LDS features.10 This suggests that full-length variant TGFBR1 and TGFBR2 proteins improperly regulate the downstream signal pathways and cause LDS1 and LDS2.
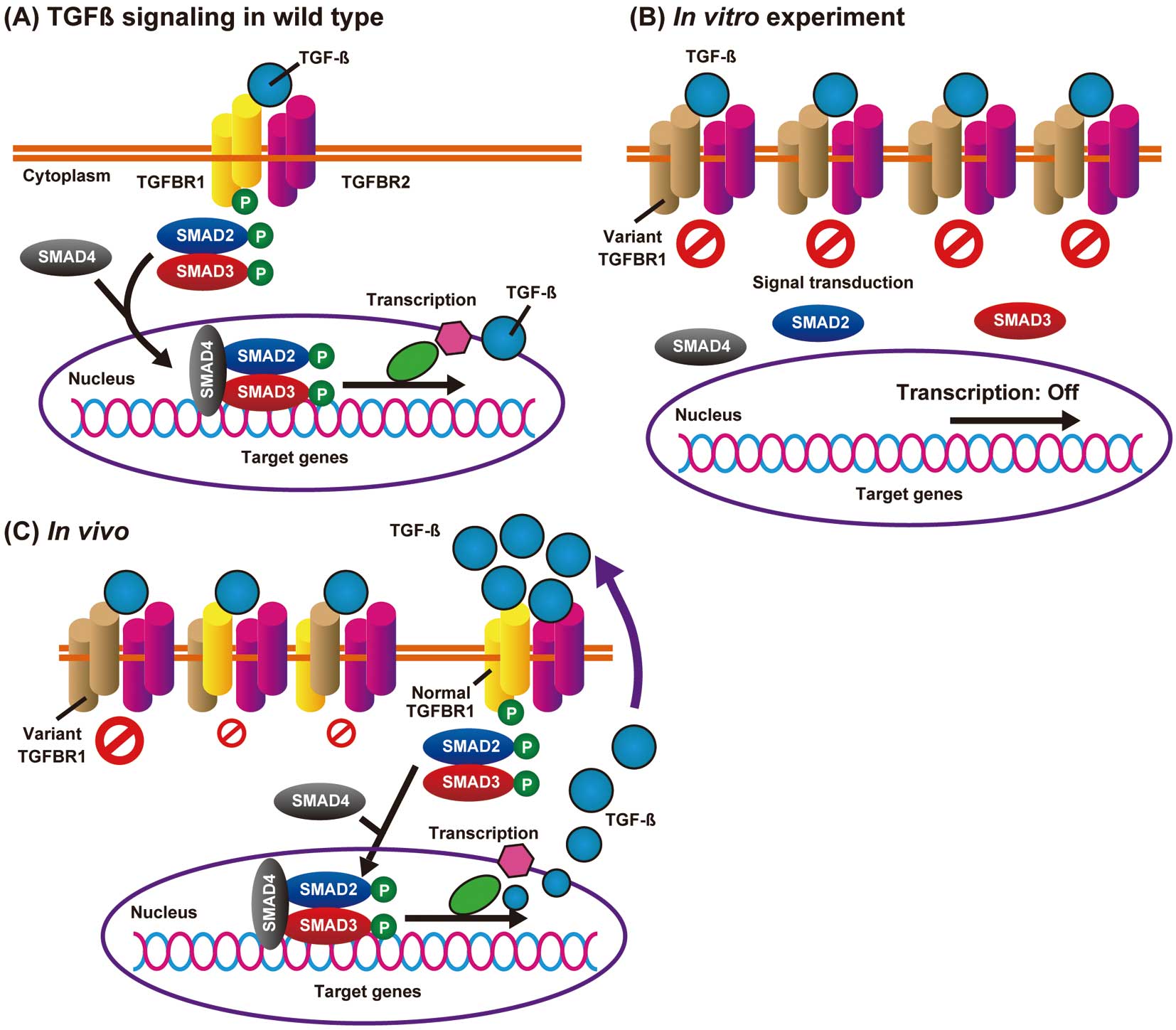
Dysregulated transforming growth factor-β (TGF-β) signaling caused by a TGF-β type I receptor (TGFBR1) missense variant. (A) In a normal aorta, TGF-β binding induces the formation of a heterotetrameric complex of two type 2 (TGFBR2; purple) and two type 1 (TGFBR1; yellow) receptors, and activates a phosphorylation cascade. (B) An in vitro functional assay using the overexpressed variant TGFBR1 (brown). The pathogenic missense variant in the serine/threonine kinase domain leads to a loss-of-function phenotype. (C) In the impaired Loeys-Dietz syndrome aortic wall, oversecreted TGF-β ligands can be theoretically transmitted through the remaining wild-type TGFBR1/TGFBR2 heterotetrameric complex.
The mechanisms of how the missense variants cause LDS1 and LDS2 are still elusive. Although most variants have been verified or predicted to cause loss of function in the in vitro cultured cells (e.g., HEK293 cells; Figure 1B),11,12 these variants enhanced TGF-β signaling in the aortic regions.10,13 The mechanism underlying the TGF-β signaling paradox remains elusive and is accepted as a scientific mystery.14 There has been speculation that increased TGF-β ligands in the LDS aortic wall activate intact TGFBR1/TGFBR2 complexes in vivo (Figure 1C), but the mechanisms of how TGF-β ligands are actively secreted remain to be determined.15
Very recently, MacFarlane et al reported a partial solution to this problem (Figure 2).16 The severely affected aortic root and ascending aorta are composed of 2 types of vascular smooth muscle cells (VSMCs): secondary heart field (SHF)- and cardiac neural crest (CNC)-derived VSMCs. In Tgfbr1M318R/+ LDS mice these 2 types of VSMCs have distinct biological properties. SHF-derived VSMCs, but not CNC-derived VSMCs, showed impaired SMAD2/SMAD3 activation in response to TGF-β, increased expression of angiotensin II (AngII) type 1 receptor (Agtr1a), enhanced responsiveness to AngII, and higher expression of TGF-β ligands. In contrast, CNC-derived VSMCs had preserved TGF-β signaling potential; and CNC-specific, but not SHF-specific, Smad2 deletion ameliorated aortic root aneurysm formation in Tgfbr1M318R/+ mice. This suggests that TGF-β ligands secreted from SHF-derived VSMCs in an AngII type I receptor (AT1R)-dependent manner could activate CNC-derived VSMCs, thereby contributing to the in vivo TGF-β overactivity in LDS aortopathy. This might also explain the usefulness of an AT1R inhibitor, losartan, for preventing aortic root aneurysm formation in LDS (Figure 2).

Model for aortic pathogenesis associated with Loeys-Dietz syndrome (LDS).16 Aortic root and ascending aorta are composed of secondary heart field (SHF)- and cardiac neural crest (CNC)-derived vascular smooth muscle cells (VSMCs). SHF- and CNC-derived VSMCs in transforming growth factor- β (TGF-β) type I receptor (Tgfbr1)M318R/+ LDS mice have different biological characteristics, and cellular responses to TGF-β are decreased and preserved in SHF- and CNC-derived VSMCs, respectively. Increased AT1R signaling stimulates extracellular signal-regulated kinase (ERK) phosphorylation and TGF-β expression in SHF-derived VSMCs. In the presence of increased TGF-β, CNC-derived cells show increased levels of phosphorylated SMAD2/SMAD3, which contributes to the pathogenesis of LDS aortopathy. Treatment with the angiotensin II type I receptor (AT1R) inhibitor losartan and CNC-specific Smad2 ablation ameliorates aortic root aneurysm formation.
As described previously, LDS1 and LDS2 are induced by loss-of-function missense variants in or near the STK domain, and full-length variant proteins seem to function improperly. Interestingly, TGFBR1 has been reported to be a causative gene for MSSE,6 and most MSSE variants are reported to be located in the extracellular ligand-binding domain, and truncating variants (nonsense, frameshift) in the STK domain. We recently encountered, however, a Japanese familial case of LDS involving a novel splice donor site variant in intron 5 in TGFBR1 (c.973+1G>A), and noted that the in silico-predicted effect of in-frame exon 5 skipping (168 bp) had the same effect as a splice acceptor site variant in intron 4 causing MSSE (c.806-2A>C; Figure 3A).6,7 Exon 5 encodes part of the STK domain, and LDS and MSSE family members do not present with overlap features; thus, we performed ex vivo minigene splicing assays of 2 variants (Figure 3B,C) to elucidate the mechanism by which these 2 apparently similar variants produce different system diseases.7

Distinct variants affecting the differential splicing of TGF-β type I receptor (TGFBR1) exon 5 cause either Loeys-Dietz syndrome (LDS) or multiple self-healing squamous epithelioma (MSSE).7 (A) Two intronic variants (c.806-2A>C and c.973+1G>A), which are both predicted to mediate in-frame exon 5 skipping, cause 2 distinct system diseases: MSSE and LDS, respectively. (B,C) Ex vivo splicing assay of 2 variants using the pTBNde(min) minigene system. (B) The c.973+1G>A LDS variant produced 2 types of in-frame products as a result of exon 5 skipping and the activation of a cryptic donor splice at a site 9 bp upstream of the 5’ natural splice donor site (arrowhead). (C) The c.806-2A>C MSSE variant activated a cryptic acceptor site at 76 bp downstream of the 3’ natural splice acceptor site (arrowhead), which produced an out-of-frame transcript and generated premature termination codon (asterisk).
On analysis of the resulting transcripts, the LDS variant was found to produce 2 types of in-frame products as a result of exon 5 skipping (r.806_973del, p.Asp269_Gln324del), and the activation of a cryptic donor splice at a site 9 bp upstream of the 5’ natural splice donor site (r.965_973del, p.Thr323_Gly325del; Figure 3B). The results were verified on reverse transcription-polymerase chain reaction using RNA from blood samples, and these 2 LDS-inducing variant proteins exerted dominant-negative effects at least in vitro. In contrast, the MSSE variant activated a cryptic acceptor site at 76 bp downstream of the 3’ natural splice acceptor site, which produced an out-of-frame transcript (r.807_882del, p. Asn270Thrfs*8), and was expected to cause haploinsufficiency because of nonsense-mediated mRNA decay (Figure 3C). Our results support the previously proposed but not yet approved mechanism6 that loss-of-function and haploinsufficiency-causing variants in the STK domain induce LDS and MSSE, respectively.
Genetic tests for suspected genetic diseases are not only for diagnosis. Variant types and sites in a single gene are known to influence the age of onset, severity, and pattern of affected organs, for example FBN1 (MFS), LMNA (laminopathy), DMD (muscular dystrophy), and TTR (hereditary amyloidogenic transthyretin amyloidosis) in the cardiovascular field, and understanding the genotype-phenotype relationship can aid in genetic counseling and in determining the treatment strategy. In contrast, to the best of our knowledge, completely distinct system and/or organ diseases induced by 1 gene mutation have been rarely reported.17
We recently reported the relationship between FBN1 genotype and severe aortic events (aortic root replacement, type A aortic dissection, and related death), and the main conclusion was that MFS patients with FBN1 truncating variants had a higher risk of aortic events compared with patients with missense and in-frame variants, and male patients had an increased risk.18 In addition, we encountered a patient with Malan syndrome with a single base insertion in exon 2 of NFIX, complicated with type A aortic dissection.19 Variants in NFIX induce 2 major types of syndrome affecting skeletal and neural development: Malan syndrome, which is characterized by Sotos-like overgrowth and macrocephaly and is mainly induced by variants affecting the evolutionarily conserved N-terminal DNA-binding/dimerization domain (exons 2 and 3); and the more severe Marshall-Smith syndrome, which is caused by frameshift and splice variants in exon 6–8 to escape nonsense-mediated RNA decay.20
Duchenne muscular dystrophy (DMD) and Becker muscular dystrophy (BMD) are neuromuscular diseases characterized by progressive muscle degeneration and weakness, and are caused by variants in the dystrophin gene (DMD) on the X chromosome. Truncating DMD variants induce the severe type of DMD, and missense or in-frame variants induce the relatively mild type of BMD.21 Variants that maintain the translational reading frame (in-frame) generally result in an abnormal but partially functional dystrophin; thus, genetic treatment approaches to restore the normal reading frame by CRISPR-Cas9 genome editing or after modifying the pre-messenger RNA splicing by “exon skipping”, especially for the more severely affected DMD male patients, have been vigorously investigated.22,23 Many patients with DMD have variable-sized deletions spanning exons 47–50; thus, an antisense oligonucleotide (eteplirsen) directed against the exon 51 splicing enhancer region of pre-mRNA, leading to its exclusion from mRNA, is approved for the treatment of DMD patients who have a confirmed variant that is amenable to exon 51 skipping (Figure 4).22

Antisense-mediated exon skipping therapy for Duchenne muscular dystrophy (DMD). (A) Schematic diagram of exon 50 deletion of DMD, causing DMD. Exons 47–50 are the deletion mutation hotspot and the variable-sized deletions produce out-of-frame transcripts that are destroyed by nonsense-mediated mRNA decay. (B) An antisense oligonucleotide compound (eteplirsen) is designed to bind to exon 51 of pre-mRNA, resulting in the exclusion of exon 51 during pre-mRNA processing to restore the reading frame. Approximately 10–15% of patients with DMD are suitable for this treatment strategy.22
Genetic tests for hereditary aortic aneurysm and dissection (HTAAD) to guide precision medicine have been covered by health insurance in Japan since 2016, and gene therapy for HTAAD has also been expected.5 There is no evidence to indicate that haploinsufficiency due to nonsense or out-of-frame variants predisposes to LDS caused by TGFBR1/TGFBR2 or to non-syndromic HTAAD caused by ATCA2 and MYH11 ; thus, gene therapy approaches to correct or destroy the affected allele could be theoretically applied to such variants. The biggest obstacle to success, however, might be a lack of established systems for the gene delivery to aortic VSMC, and various delivery methods are being developed.24,25
We briefly reviewed the recent understanding of the molecular mechanism of LDS and the unresolved problems, including the TGF-β signaling paradox and the mechanism by which TGFBR1 variants cause 2 distinct system diseases, LDS and MSSE. Recent basic research using the LDS mice model provides crucial insights into the pathogenic mechanisms of LDS, and the establishment of an integrated clinical and genomic information system for genetic diseases would also contribute to the discovery of promising clues to the mechanism behind gene mutation.9 Further analysis is also warranted to analyze genetic variants of unknown clinical and biological significance, as in the present study, to deepen the understanding of the disease mechanism.
I.K. is a member of Circulation Reports ’ Editorial Team. T.F. is affiliated with an endowed department sponsored by Actelion Pharmaceuticals Japan, Otsuka Pharmaceutical, Nipro Corporation, Terumo Corporation, Senko Medical Instrument, Century Medical, KCI Licensing, Abbott Medical Japan, Mochida Pharmaceutical, Nippon Shinyaku and Teijin Pharma Limited. The other authors declare no conflicts of interest.