2014 Volume 39 Issue 2 Pages 93-100
2014 Volume 39 Issue 2 Pages 93-100
In meiosis, pairing and recombination of homologous chromosomes are crucial for the correct segregation of chromosomes, and substantial movements of chromosomes are required to achieve homolog pairing. During this process, it is known that telomeres cluster to form a bouquet arrangement of chromosomes. The fission yeast Schizosaccharomyces pombe provides a striking example of bouquet formation, after which the entire nucleus oscillates between the cell poles (these oscillations are generally called horsetail nuclear movements) while the telomeres remain clustered to the spindle pole body (SPB; a centrosome-equivalent structure in fungi) at the leading edge of the moving nucleus. S. pombe mutants defective in telomere clustering frequently form aberrant spindles, such as monopolar or nonpolar spindles, leading to missegregation of the chromosomes at the subsequent meiotic divisions. Here we demonstrate that such defects in meiotic spindle formation caused by loss of meiotic telomere clustering are rescued when nuclear movement is prevented. On the other hand, stopping nuclear movement does not rescue defects in telomere clustering, nor chromosome missgregation even in cells that have formed a bipolar spindle. These results suggest that movement of the SPB without attachment of telomeres leads to the formation of aberrant spindles, but that recovering bipolar spindles is not sufficient for rescue of chromosome missegregation in mutants lacking telomere clustering.
In meiosis, one round of DNA replication is followed by two consecutive rounds of chromosome segregation to generate four haploid gametes from a parental diploid cell. During this process, recombination between homologous chromosomes occurs to generate a recombined set of the haploid genome. In a wide variety of organisms, telomeres form a cluster adjacent to the nuclear envelope in meiotic prophase (reviewed in Scherthan, 2001; Hiraoka and Dernburg, 2009). It has been shown that telomere clustering plays a role in recombination of homologous chromosomes, which in turn is crucial for the proper segregation of chromosomes. The most striking example of telomere clustering in meiotic prophase is observed in the fission yeast S. pombe. In this organism, during meiotic prophase, the centromeres detach from the spindle pole body (SPB; a centrosome-equivalent structure in fungi) and the telomeres cluster next to the SPB (Chikashige et al., 1994). Subsequently, the reorganized nucleus elongates and oscillates between the cell poles (and in this state is referred to as the horsetail nucleus) in meiotic prophase, and the telomeres remain clustered near the SPB during the nuclear movement. Nuclear movement is driven by cytoplasmic microtubules through their interaction with clustered telomeres across the nuclear envelope (Chikashige et al., 2006, 2007; Hiraoka and Dernburg, 2009). Upon entering meiotic divisions, the SPB leading the nuclear movement stops around the center of the cell and cytoplasmic microtubules are reorganized into spindle microtubules for chromosome segregation (Ding et al., 1998; Yamamoto et al., 2001).
Lines of evidence suggest that formation of meiotic spindles involves the characteristic behaviors of telomeres that are unique to meiosis (Tomita and Cooper, 2007; Ohta et al., 2012; Tomita et al., 2013). It has been shown that loss of telomere clustering in meiotic prophase correlates with defects in spindle formation in the subsequent meiotic division: formation of aberrant meiotic spindles has been observed in mutants defective in telomere clustering such as the bqt1Δ, taz1Δ, and rap1Δ mutants (Tomita and Cooper, 2007), the bqt4Δ mutant (Chikashige et al., 2009), and also the bqt2Δ mutant (confirmed in this report). Here we demonstrate that such defects in meiotic spindle formation in bqt2Δ cells are rescued when nuclear movement is prevented. On the other hand, stopping nuclear movement does not rescue chromosome missgregation even in cells that have formed a bipolar spindle in the absence of telomere clustering. Thus, in the absence of telomere clustering, chromosome missgregation occurs independently of spindle defects.
The genotypes of the strains used in this paper are shown in Supplemental Table S1. YE, YES, or EMM2 were used for routine mitotic culture of S. pombe cells, and ME agar plates were used to induce meiosis of h90 cells. The chemical compositions of YE, YES, EMM2 and ME media are described in Moreno et al. (1991). Induction of meiosis has been described previously (Chikashige et al., 2006).
Fluorescent fusion constructsGFP (or CFP)-Atb2 fusion constructs were made as follows: The nda3 promoter, the coding sequence of the GFP-S65T or CFP gene, the coding sequence of the atb2+ gene, and the nmt1 terminator sequence were ligated into the integration vector pYC36 (Chikashige et al., 2004). The resulting plasmid was integrated into the chromosome at the lys1 gene locus. The mCherry-Bqt4 fusion constructs were made as follows: the coding sequence of the mCherry gene was ligated in-frame between the bqt4 promoter sequence and the bqt4 coding sequence with the nmt1 terminator sequence, and it was integrated into the chromosome at the aur1 gene locus using the aur1R allele, which dominantly confers resistance to Aureobasidin A (Takara) (Hashida-Okado et al., 1998). Strains carrying Sid4-GFP, Sid4-mRFP, Taz1-mCherry and Hht1-mRFP were constructed by replacing each wild-type gene with the selection marker kanr by a PCR-based gene targeting method (Bähler et al., 1998).
Image acquisition and processingFluorescence microscope images were obtained using a computer-controlled fluorescence microscope system (DeltaVision; Applied Precision, Inc., Seattle, WA). For imaging of live cells, a DeltaVision microscope system set up in a temperature-controlled room was used (Haraguchi et al., 1999). This microscope system is based on an inverted fluorescence microscope (IX70, Olympus Optical) equipped with a charge-coupled device (CoolSNAP HQ, Photometrics, Tuson, AZ). An Olympus oil immersion objective lens (Plan Apo 60X, NA=1.4) was used for observation. For time-lapse observation, living cells were mounted in a 35 mm glass-bottom culture dish (MatTek Corp., Ashland, MA) coated with lectin or a microfluidic flow chamber for yeast (CELLASIC Y04C), and observed in EMM2-N medium at 26°C for meiotic cells. Images were acquired using SoftWoRx software, provided as part of the DeltaVision system. A 3D stack of images spanning 9–15 focal planes at 0.3 μm increments was recorded at each time point. Images in Fig. 2 were processed by the denoising algorithm (Boulanger et al., 2009), and subsequently by constrained iterative deconvolution (Agard et al., 1989). This sequence of processing has been shown to greatly improve fluorescent signals while keeping the object shapes virtually intact (Matsuda et al., 2010). Projection images were generated using a maximum intensity method.
Quantification of horsetail nuclear movementSPBs and chromosomes were visualized with Sid4-GFP and Hht1-mRFP, respectively. Projections of optical sections at each time point were used in the following analysis. The coordinates of the Sid4-GFP foci were automatically acquired by our custom program based on a least-squares fitting of a Gaussian profile to the fluorescence intensity. To confirm the coordinates, the trajectory of the SPB was visually checked by plotting the coordinates on images through all time points. Displacements of the Sid4-GFP coordinates for each of the 30-sec intervals were summed for the 21 time points corresponding to the 10-minute period from 65 minutes to 55 minutes before the first meiotic division.
Following the spindle microtubules during meiosis in living cells of S. pombe, we confirmed that aberrant meiotic spindles were formed in bqt2Δ mutant cells defective in telomere clustering, as previously reported in the bqt1Δ, taz1Δ, and rap1Δ mutants (Tomita and Cooper, 2007) as well as in the bqt4Δ mutant (Chikashige et al., 2009). Fig. 1 shows an example of bqt2Δ mutant cells. In the first meiotic division of this mutant, bipolar spindles were observed only in 17 out of 45 cells (38%); monopolar spindles were observed in 16 out of 45 cells (36%) and spindles with no poles (nonpolar spindles) in 12 out of 45 cells (27%), as summarized in Fig. 4C. In wild-type cells, monopolar or nonpolar spindles were not observed (Fig. 4C). Essentially the same results were reproduced in independent experiments as summarized in Supplemental Table S2 (also see Fig. 6E). The frequency of spindle defects was measured in living zygotes that proceeded with nuclear fusion through the first meiotic division; zygotes displaying failures in nuclear fusion were excluded.

Defects in meiotic spindle formation in bqt2Δ cells. (A) An example of a bipolar spindle in meiosis I (38%; 17 of 45 cells examined) (B): An example of a V-shaped monopolar spindle in meiosis I (36%; 16 of 45 cells examined). (C): An example of a nonpolar spindle where the first meiotic spindle lacks Sid4 signals at both ends (27%; 12 of 45 cells examined). Bar indicates 10 μm. Time-lapse images were recorded at intervals of 5 minutes. Images at selected time points are shown; numbers on the left of the images indicate the time in minutes (time 0 is the point at which the spindles of the first meiotic division appear). The image at each time point is a projection of optical section images. Enlarged images of the regions indicated by the red boxes in (A), (B), and (C) are shown in (D), (E), and (F), respectively. The observed strain is CRLw89 (bqt2Δ). Tubulin (green) and SPBs (magenta) were visualized using GFP-Atb2 and Sid4-mRFP, respectively.
Disengagement of the SPB from the nuclear envelope in mutant cells defective in telomere clustering has been reported (Tomita and Cooper, 2007). To elucidate causes for the occurrence of monopolar or nonpolar spindles, we followed movements of the SPB relative to the nuclear envelope in living cells. An example of monopolar spindles is shown in Fig. 2A and B (also see Supplemental Movie 1). In this example, a monopolar V-shaped spindle was formed (25 min); the monopolar spindle elongated during meiosis I, causing an aberrant nuclear division (50–70 min); and by the end of this period, a fragment of the SPB had disengaged from the nucleus (70 min). During meiosis II, the SPB remaining associated with the nucleus formed a normal bipolar spindle, whereas the disengaged SPB did not (85–110 min). An example of nonpolar spindles is shown in Fig. 2C and D (also see Supplemental Movie 2). In this example, the entire SPB was disengaged from the nucleus (0 min) and the disengaged SPB did not nucleate spindle microtubules. Nevertheless, a nonpolar spindle was formed, with no SPBs at either end, at meiosis I (20 min) and meiosis II (110 min) and this was followed by the generation of nuclear fragments. These observations suggest that monopolar or nonpolar spindles result from disengagement of the SPB, or a part of the SPB, from the nucleus.

Disengagement of the SPB from the nucleus in bqt2Δ cells. (A, C) Time-lapse images of the bqt2Δ mutant (CRLz05) were recorded at 5 minute intervals. Movies for (A) and (C) can be seen in Supplemental Movie 1 and Supplemental Movie 2, respectively. Images at selected time points are shown; the numbers on the left of the images indicate the time in minutes (at time 0 the spindles of the first meiotic division appear). The image at each time point is a projection of optical section images. Bar indicates 10 μm. (B) and (D) are selected frames taken at the times indicated by the red boxes in (A) and (C), respectively. Tubulin (green, “Tub”), the SPB (red), and the nuclear envelope (blue, “NE”) were visualized using CFP-Atb2, Sid4-GFP and mCherry-Bqt4, respectively. In this set of experiments, the occurrence of monopolar and nonpolar spindles at meiosis I was 24% (14 out of 58 cells) and 17% (10 out of 58 cells), respectively.
Conceivably, defects in the formation of the bipolar spindle could be caused by horsetail nuclear movement in the absence of telomere-SPB attachment. To test this possibility, we examined spindle formation in the bqt2Δ mutant combined with mutations conferring defective nuclear movements (Fig. 3). Hrs1 (also called Mcp6) is a meiosis-specific component of the SPB, and hrs1Δ mutant cells show defects in nuclear movement (Fig. 3A, B) as previously reported (Tanaka et al., 2005; Saito et al., 2005). Cells of the dhc1Δ (dynein heavy chain) mutant also show defective nuclear movement (Fig. 3B) (Yamamoto et al., 1999). In hrs1Δ mutant cells, meiotic bipolar spindles formed normally (Fig. 4A, B). Moreover, the frequency of the bipolar spindles increased from 38% in bqt2Δ single mutant cells to about 90% in bqt2Δ hrs1Δ double mutant cells (Fig. 4C). A similar effect was observed in the dhc1Δ mutant: the spindle defects in bqt2Δ cells were rescued by dhc1 deletion (Fig. 4C). These results were reproduced in independent experiments shown in Fig. 6 (summarized in Supplemental Table S2). Hence, the spindle defects caused by the loss of telomere clustering were rescued by stopping nuclear movement. Mechanical forces engaged on the cytoplasmic side of the SPB during horsetail nuclear movement may need to be balanced by chromosomes attached to the nucleoplasmic side of the SPB. We speculate that chromosomes rein back the SPB, providing a buffer against the cytoplasmic forces generated by horsetail nuclear movements.

Quantification of horsetail nuclear movements. (A) Time-lapse images were recorded at 30 second intervals. The image at each time point is a projection of optical section images. Chromosomes (magenta) and the SPB (green) were visualized by Hht1(histone H3)-mRFP and Sid4-GFP, respectively. Numbers on the left of the images indicate the time in minutes (where time 0 corresponds to division of the SPB at the first meiotic division). The bar indicates 10 μm. (B) The total distance of the SPB movements during the 10 minutes from –65 minutes to –55 minutes were calculated as described in the Materials and Methods. The strains used here were CRL065 (wild type), CRL03g (bqt2Δ), CRL063 (hrs1Δ), CRL03f (bqt2Δ hrs1Δ), CRLy35 (dhc1Δ) and CRLy37 (bqt2Δ dhc1Δ). The number of cells examined is shown in the parentheses below each graph. The error bar represents the standard deviation.
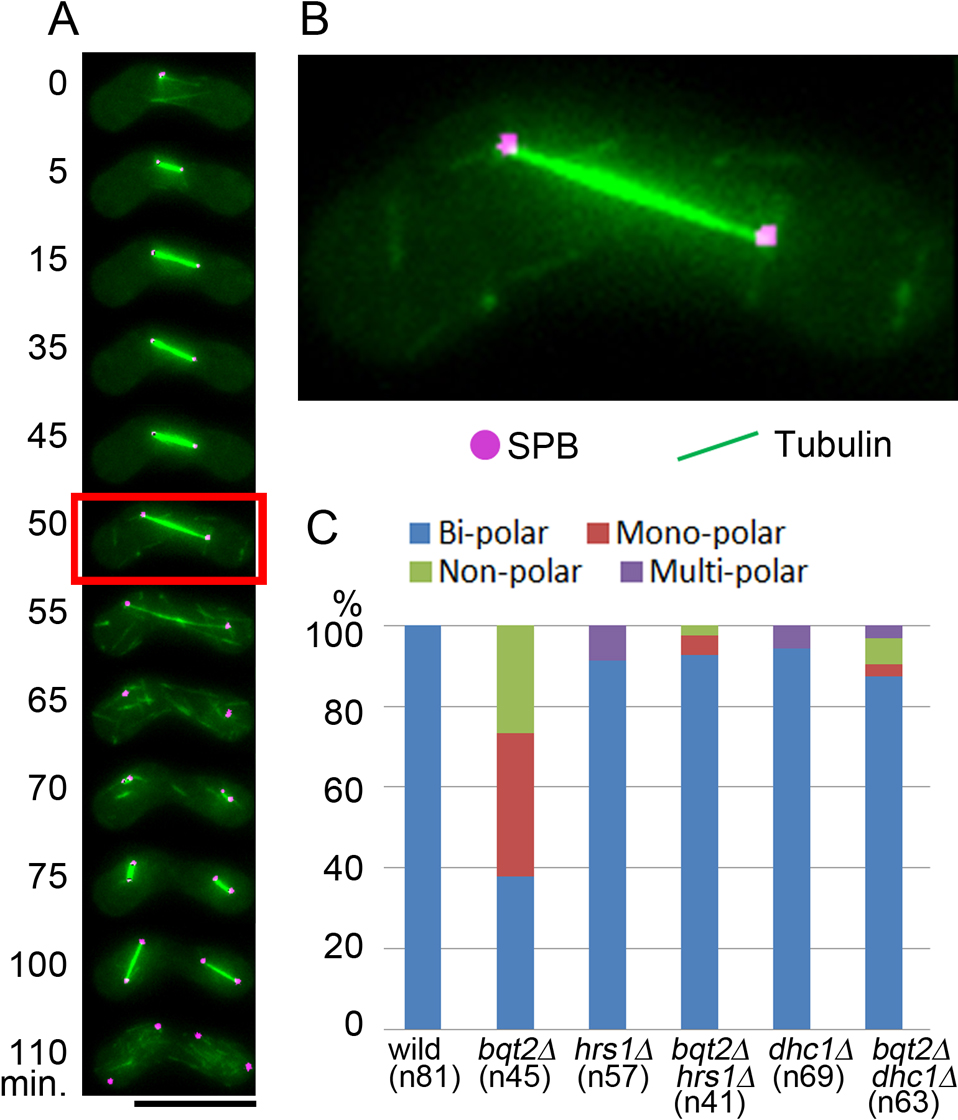
The spindle defects in bqt2Δ cells are rescued by stopping nuclear movement. (A) An example of live-imaging of the SPB (magenta) and microtubules (green) in the bqt2Δ hrs1Δ double mutant (CRLx99). The numbers on the left of the images indicate the time in minutes (time 0 is defined as the frame at which the spindle first appears at the first meiotic division). The bar indicates 10 μm. (B) The bipolar spindle in bqt2Δ hrs1Δ cells was seen at the 50 min time point, indicated by a red box in (A). (C) The frequencies of bipolar, monopolar, nonpolar and multipolar spindles in the strains are indicated. The frequency of monopolar and nonpolar spindles decreased and the frequency of bipolar spindles increased in both the bqt2Δ hrs1Δ and bqt2Δ dhc1Δ cells. The observed strains were CRLy31 (wild type), CRLw89 (bqt2Δ), CRLy70 (hrs1Δ), CRLx99 (bqt2Δ hrs1Δ), CRLy35 (dhc1Δ) and CRLy37 (bqt2Δ dhc1Δ). The number of cells examined is shown in the parentheses below each graph.
These results raise an apparent discrepancy with a previous report in which dhc1∆ bqt1∆ zygotes suffered defective spindle formation reminiscent of bqt1∆ single mutant zygotes (Tomita and Cooper, 2007). It should be pointed out that dhc1∆ bqt2∆ cells sometimes cause failures in nuclear fusion. In our analysis, we followed living zygotes from nuclear fusion to the first meiotic division, and excluded zygotes displaying failures in nuclear fusion. Such zygotes were included in the previous analyses; re-examination of these analyses excluding all dhc1∆ bqt1∆ zygotes displaying nuclear fusion defects, found that the majority of dhc1∆ bqt1∆ zygotes show proper bipolar spindle formation (J. P. Cooper, personal communication). Thus, the discrepancy can be reconciled by counting only zygotes that completed nuclear fusion.
Chromosome segregation defects in telomere-defective mutants are not rescued by stopping nuclear movementBecause spindle defects in bqt2Δ cells were rescued by hrs1Δ, we next examined whether telomere clustering is recovered in the bqt2Δ hrs1Δ mutant strain. Fig. 5A shows the telomeres, SPB and microtubules in a living zygote of the bqt2Δ hrs1Δ strain: the telomeres are diffused from the SPB (Fig. 5B) while the bipolar spindle is formed (Fig. 5C) in this example. Whereas the telomeres formed a single cluster in all of the examined wild-type cells (26 out of 26 cells) and almost all of the hrs1Δ cells (52 out of 53 cells), fully clustered telomeres were never observed in the examined bqt2Δ and bqt2Δ hrs1Δ cells (54 bqt2Δ and 37 bqt2Δ hrs1Δ cells). Thus, we conclude that defects in telomere clustering are not rescued by preventing nuclear movement.

Telomere clustering defects in bqt2Δ cells are not rescued by hrs1 deletion. (A) An example of live-cell imaging of microtubules (blue; CFP-Atb2), the SPB (red; Sid4-GFP) and telomeres (green; Taz1-mCherry) in the bqt2Δ hrs1Δ double mutant (CRLz41). The numbers on the left of the images indicate the time points in minutes (where time 0 corresponds to the first appearance of the spindle at the first meiotic division). (B) A magnification of the region indicated by the red box in (A), showing that telomeres (indicated by white arrows) were scattered in the horsetail nucleus in the bqt2Δ hrs1Δ double mutant at the –40 min time point. The bar indicates 10 μm. (C) A magnification of the region indicated by the red box in (A), showing the bipolar spindle seen at the 45 min time point. The bar indicates 10 μm. The strains used here to examine telomere clustering were CRLy51 (wild type), CRLy50 (bqt2Δ), CRLy44 (hrs1Δ) and CRLy52, CRLz41 (bqt2Δ hrs1Δ).
Next we observed chromosome segregation in living cells of the mutants. Chromosome segregation patterns were examined in cells that produced the bipolar spindle to eliminate the effects of gross spindle defects. Aberrant segregation of chromosomes was frequently observed in bqt2Δ hrs1Δ cells even in the presence of the bipolar spindle. The example in Fig. 6A and B shows unequal segregation of the chromosomes producing two divided nuclei of unequal size and fluorescence intensity. Another example, in Fig. 6C and D, shows a lagging chromosome left behind divided nuclei. To quantify the frequency of chromosome segregation errors separately from spindle defects, the incidence of unequal segregation and lagging chromosomes in cells that had produced a normal bipolar spindle was determined. In this set of experiments, the frequencies of bipolar, monopolar and nonpolar spindles shown in Fig. 6E were similar to those shown in Fig. 4C (summarized in Supplemental Table S2). The frequency of aberrant chromosome segregation in bqt2Δ hrs1Δ cells was comparable to that in bqt2Δ cells, indicating that chromosome segregation errors in bqt2Δ cells are not rescued by preventing nuclear movement.

Chromosome segregation defects in bqt2Δ hrs1Δ cells. (A, C) Time-lapse images of the bqt2Δ hrs1Δ double mutant (CRL03q) were recorded at 5 minute intervals. The image at each time point is a projection of optical section images. The numbers on the left of the images indicate the time in minutes (time 0 corresponds to the first appearance of spindles in the first meiotic division). (B) and (D) show selected frames taken at the times indicated by the red boxes in (A) and (C), respectively. Tubulin (green, “Tub”), the SPB (red), and chromosomes (blue, “Chr”) were visualized using CFP-Atb2, Sid4-GFP and Hht1(histone H3)-mRFP, respectively. The bar indicates 10 μm. (A, B) Examples of unequal chromosome distribution at the first division. (C, D) Examples of lagging chromosomes (arrow in D) at the first division. (E) Frequencies of bipolar, monopolar, nonpolar and multipolar spindles in the strains were as indicated. The observed strains were CRL02t (wild type), CRL03r (bqt2Δ), CRLo11a (hrs1Δ), and CRL03q (bqt2Δ hrs1Δ). The number of cells examined is shown in the parentheses below each graph. In this set of experiments, the occurrence of bipolar spindles at meiosis I was 89 out of 89 wild-type cells, 26 out of 81 bqt2Δ cells, 79 out of 84 hrs1Δ cells, and 72 out of 84 bqt2Δ hrs1Δ cells. (F) The frequencies of equal and unequal chromosome distribution, and of lagging chromosomes at the first division in the cells that were observed to form a bipolar spindle. The number of cells that formed a bipolar spindle at the first division is shown in the parentheses below each graph.
A previous study proposed that SPB maturation may depend on telomere clustering and that aberrant spindles caused by defects in telomere clustering could contribute to chromosome missegregation (Tomita and Cooper, 2007). Our results do not eliminate the possibility that telomere-dependent SPB maturation may exist, but argue against spindle defects as a major cause of chromosome missegregation in bouquet-defective backgrounds, as stopping horsetail nuclear movement can circumvent the aberrant spindle formation but cannot rescue chromosome missegregation. Because telomeres are diffused in bouquet mutants when horsetail nuclear movement is prevented and chromosome missegregation occurs even when a normal bipolar spindle is formed in those cells, defective telomere clustering could be a direct cause of chromosome missegregation that is independent of spindle defects. It is likely that chromosome missegregation in the absence of telomere clustering can be attributed to decreased levels of homologous recombination. Thus, mutants defective in telomere clustering that do not exhibit nuclear movement (for example, the bqt2Δ hrs1Δ double mutant) separate the direct effects of the loss of telomere clustering from the secondary effects of nuclear movement, and provide an opportunity to determine the significance of telomere clustering.
We thank Julie Cooper for providing the S. pombe strain expressing Hht1-mRFP, for sharing unpublished results as personal communication, and for critically reading the manuscript. This work was supported by grants KAKENHI (22370074 to YC, 24770113 to AM, 25116006 to TH, and 26251037 to YH).