2019 Volume 44 Issue 2 Pages 85-94
2019 Volume 44 Issue 2 Pages 85-94
In research on cell biology, organelles have been a major unit of such analyses. Researchers have assumed that the inside of an organelle is almost uniform in regards to its function, even though each organelle has multiple functions. However, we are now facing conundrums that cannot be resolved so long as we regard organelles as functionally uniform units. For instance, how can cells control the diverse patterns of glycosylation of various secretory proteins in the endoplasmic reticulum and Golgi in an orderly manner with high accuracy? Here, we introduce the novel concept of organelle zones as a solution; each organelle has functionally distinct zones, and zones in different organelles closely interact each other in order to perform complex cellular functions. This Copernican Revolution from organelle biology to organelle zone biology will drastically change and advance our thoughts about cells.
Key words: organelle zone, contact site, ER stress, Golgi stress, organelle autoregulation
Eukaryotic cells contain various organelles, such as the nucleus, mitochondria, endoplasmic reticulum (ER), Golgi apparatus, lysosomes, and peroxisomes. The main functions of these organelles are the storage of genetic information, ATP synthesis, synthesis of secretory proteins, post-translational modification of secretory proteins, degradation of macromolecules, and β-oxidation of long chain fatty acids, respectively. However, it is well known that each organelle has various functions.
In conventional cell biology, we have analyzed each organelle as a unit. With the development of super-resolution microscopes such as STED (stimulated emission depletion), PALM (photo activated localization microscopy), STORM (Stochastic optical reconstruction microscopy) and SCLIM (super-resolution con- focal live imaging microscopy) (Nakano, 2013), we have realized that each organelle can be divided into multiple regions that perform distinct functions. These functionally distinct regions in an organelle are called “organelle zones” (Nishitoh, 2019; Shimizu, 2019; Tamura et al., 2019). The novel concept of an organelle zone is essential to understand each distinct function of organelles. In other words, cell biology has just encountered a Copernican Revolution: from organelle biology to organelle zone biology (Fig. 1).
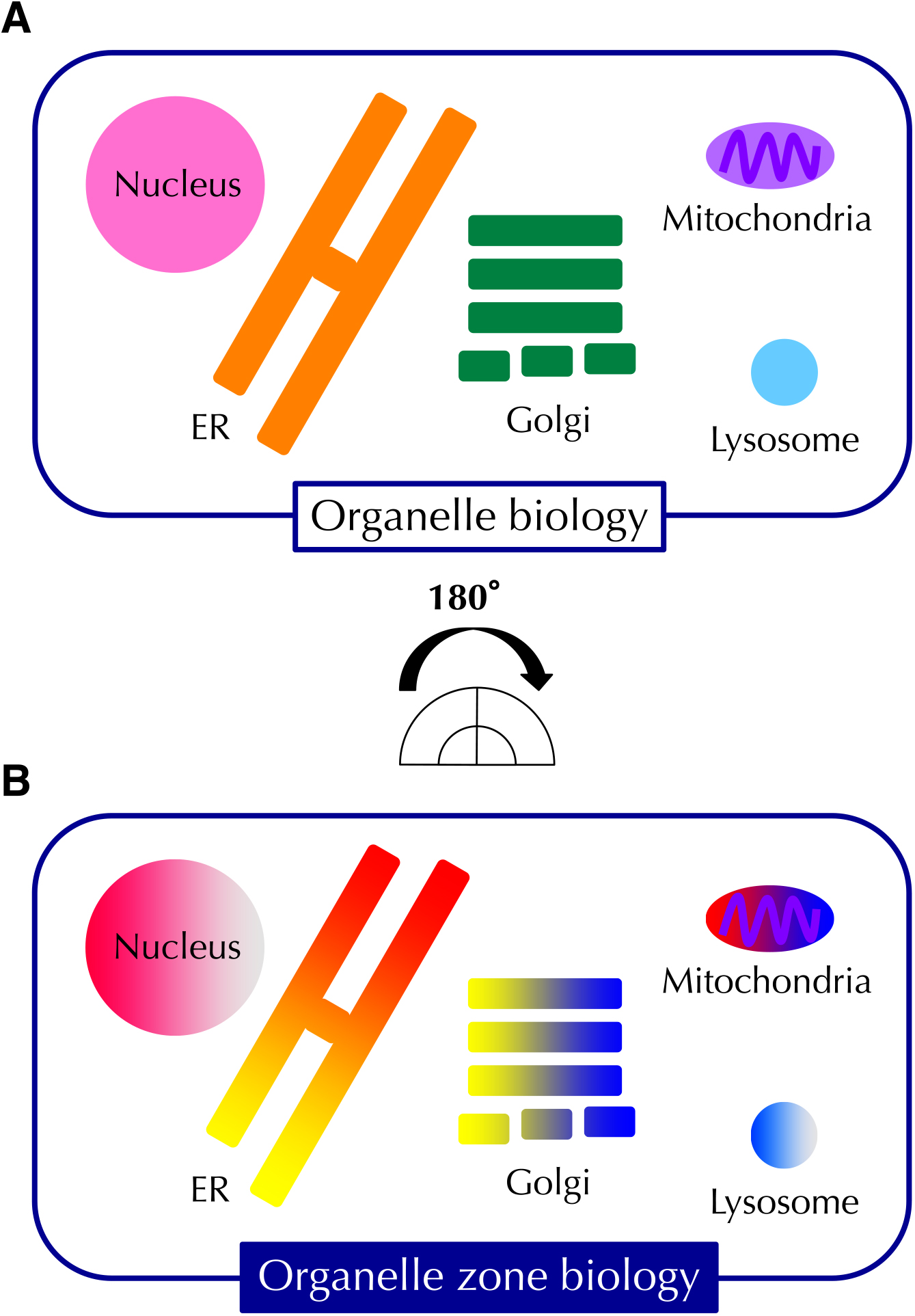
From organelle biology to organelle zone biology. (A) Schematic representation of the traditional concept of “organelle biology with the inside of organelles thought to be uniform. (B) Schematic of the novel concept of “organelle zone biology, where each organelle contains distinct zones that are specialized for specific functions.
It has been a mystery how different proteins receive distinct post-translational modifications in the Golgi apparatus. To clarify this fundamental issue in cell biology, Dr. Satoshi Goto analyzed the localization of membrane proteins in the wing disc of fruit fly, and found that FRINGE-CONNECTION (FRC), SULFATELESS (SFL), and RHOMBOID (RHO) receive post-translational modifications in distinct Golgi compartments from each other, and he proposed that different proteins can receive their correct modification because of the separate localization of the modification enzymes and their substrates in distinct zones (Yano et al., 2005) (Fig. 2). After this epoch-making discovery, various organelle zones has been discovered, leading to the establishment of the novel concept of organelle zones. Without this concept, it is impossible to understand the function of organelles, because now we know that organelles perform a number of functions without entanglement.
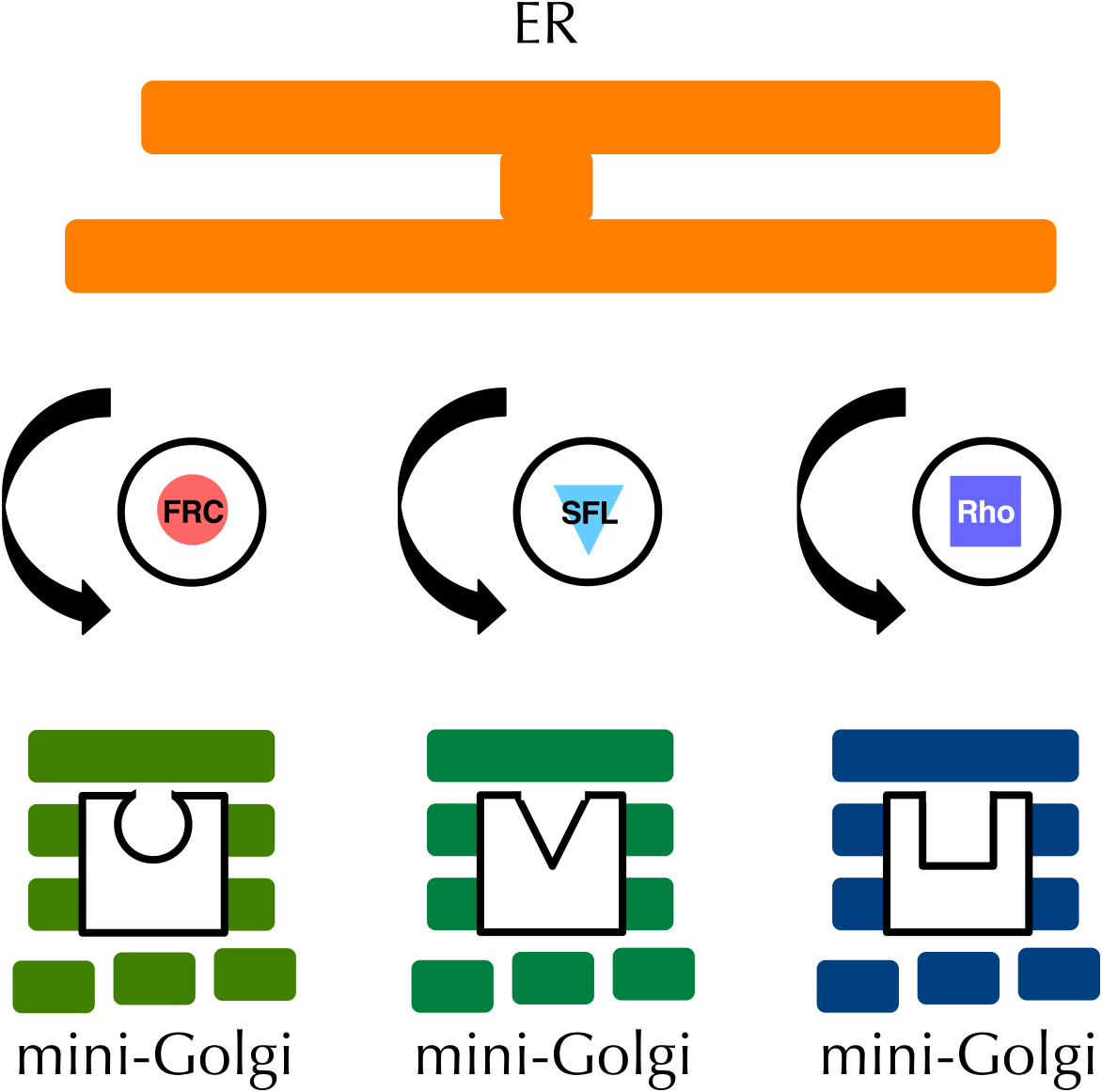
Discovery of organelle zones. In the imaginal discs of Drosophila melanogaster, proteins such as FRC, SFL, and rhomboid are localized in distinct mini-Golgi and receive different post-translational modifications.
Because research into organelle zones has been developing exponentially, it seems impossible to describe all organelle zones. Therefore, this review will concisely describe well-known organelle zones.
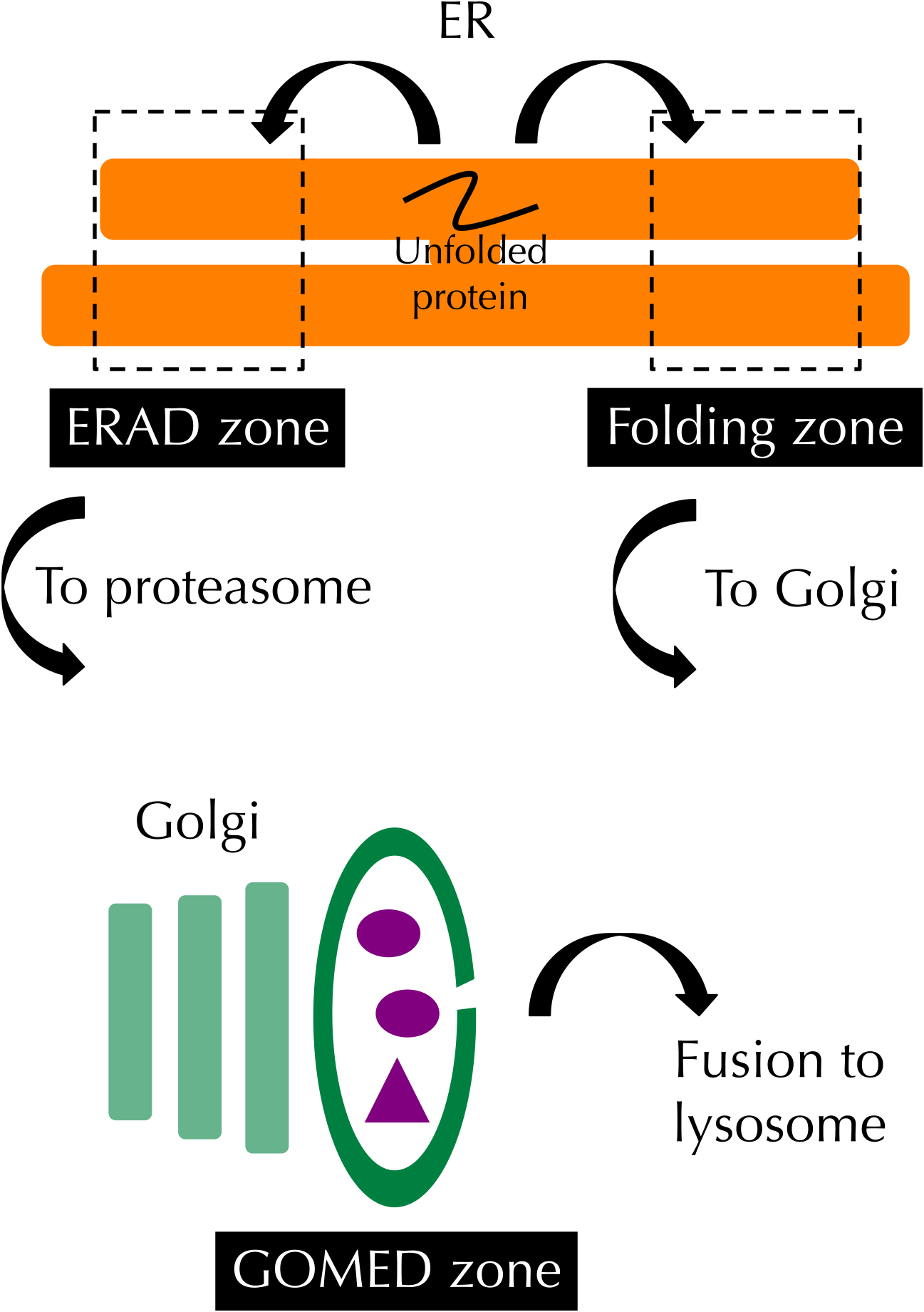
Organelle zones for quality control of proteins in the ER and Golgi. ER chaperones are localized in the folding zone in the ER and assist with the folding of secretory and membrane proteins synthesized in the ER. In the ERAD zone, the components of ERAD accumulate and promote the retrotransportation of unfolded or malfolded proteins from the ER to the cytosol to enhance their degradation by the proteasome. Some parts of the Golgi membrane are recruited for ATG5-independent autophagy, termed GOMED.
The ER is the organelle where secretory and membrane proteins are synthesized and folded with the assistance of ER chaperones such as BiP and calnexin. Proteins that cannot be folded correctly are degraded by a mechanism called ER-associated degradation (ERAD). Lederkremer and colleagues reported that ER α1,2 mannosidase I (ERManI), a central component of ERAD, is located in an ER-derived quality control compartment (also called quality control vesicles), to which ERAD substrates are transported and where they interact with the enzyme (Avezov et al., 2008; Benyair et al., 2015). Their observations suggested that the ER contains distinct functional zones, namely a folding zone and an ERAD zone.
Shimizu and colleagues reported the Golgi membrane-associated degradation (GOMED) zone for quality control of proteins in the Golgi (Yamaguchi et al., 2016). Insulin is a hormone to reduce the level of blood glucose. When the glucose level decreases enough, insulin stored in secretory vesicles in pancreatic β cells is degraded by unconventional autophagy derived from specific trans-Golgi membranes in an ATG5-independent manner. This mechanism is called GOMED.
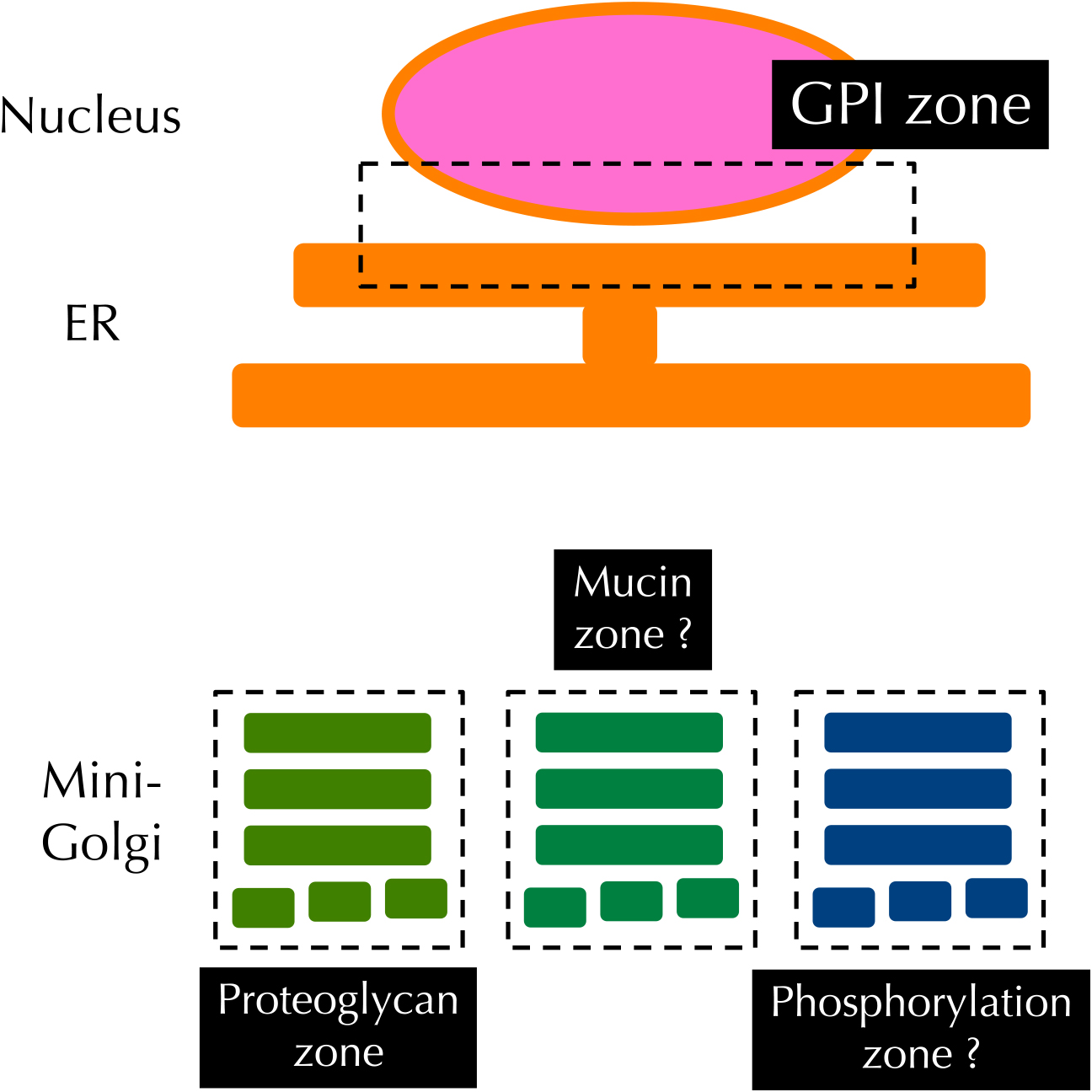
Organelle zones responsible for post-translational modification in the ER and Golgi. Enzymes involved in the post-translational modification of GPI-anchored proteins are localized in part of the ER (the nuclear envelope), whereas those required for proteoglycan glycosylation are located in the proteoglycan zone of the Golgi apparatus. Proteins important for phosphorylation and mucin-glycosylation are speculated to be localized in the mucin zone and the phosphorylation zone.
Secretory and membrane proteins receive distinct post-translational modifications in the ER and Golgi after their synthesis in the ER. It had been unclear how cells control the specificity of modification in order to allow each protein to receive its specific modification correctly. Recent studies have revealed that each post-translational modification in the ER and Golgi occurs in separate zones that are physically separated in order to ensure substrate specificity.
(1) GPI zoneGlycosylphosphatidylinositol (GPI)-anchored proteins are membrane proteins that are anchored in the membrane through a GPI motif. Maturation of GPI-anchored proteins is very complex and consists of three steps (Ferguson et al., 2015). The first step is synthesis of GPI on the ER membrane, and the second step is transfer of GPI to proteins, whereas the third step is remodeling of the lipid chain in the ER and Golgi. Goto and colleagues revealed that PIG-B and PIG-O (enzymes for step 1) as well as PIG-T (an enzyme for step 2) and mRNA of Dlp (a GPI-anchored protein) were localized in a limited region of the ER membrane close to the nucleus (GPI zone) (Yamamoto-Hino et al., 2018), suggesting that the synthesis and glycosylation of GPI-anchored proteins occurs in the GPI zone of Drosophila culture cells. When PIG-G was artificially expressed outside the GPI zone but still in the ER, the efficiency of GPI glycosylation was markedly reduced, suggesting that formation of the GPI zone is essential for the maturation of GPI-anchored proteins. It is still unclear how PIG enzymes and their substrates are localized in the GPI zone.
(2) Proteoglycan zoneProteoglycans are glycoproteins containing glycosaminoglycans (GAGs) (Nadanaka and Kitagawa, 2008). Core proteins of proteoglycans are synthesized in the ER and receive post-translational modifications such as glycosylation and sulfation in the Golgi. In fruit fly, Drosophila melanogaster, the Golgi apparatus forms a number of mini-Golgi in the cell, which all have cis-, medial-, and trans-Golgi cisternae. Goto and colleagues found that SFL, the sulfation enzyme for GAG, is localized to a subset of the mini Golgi distinct from those harboring the UDP-sugar transporter FRC, and from those with the protease RHO, which cleaves the glycoprotein SPITZ (SPI) in the wing disc of fruit fly (Yano et al., 2005) (Fig. 2). This suggested an interesting possibility that the maturation of proteoglycans occurs in specific Golgi stacks, namely, the proteoglycan zone.
(3) Mucin zone and phosphorylation zone in the GolgiMucins are another type of glycoprotein distinct from proteoglycans (Jensen et al., 2010). Similar to proteoglycans, the core proteins of mucins receive mucin-type of glycosylation in the Golgi. The glycosylation process of mucin is very complex and a number of enzymes are involved in this process. To avoid inappropriate glycosylation between proteoglycans and mucins, cells may have a zone specialized for mucin glycosylation (the mucin zone), although this has not yet been identified experimentally.
Four-jointed is a kinase localized in the Golgi apparatus that phosphorylates luminal domains of various substrates in fruit fly (Ishikawa et al., 2008). Ishikawa and colleagues identified a phosphorylation zone in the Golgi apparatus, in which Four-jointed is localized (Ishikawa et al., 2008), which may be important for the phosphorylation of specific substrates.
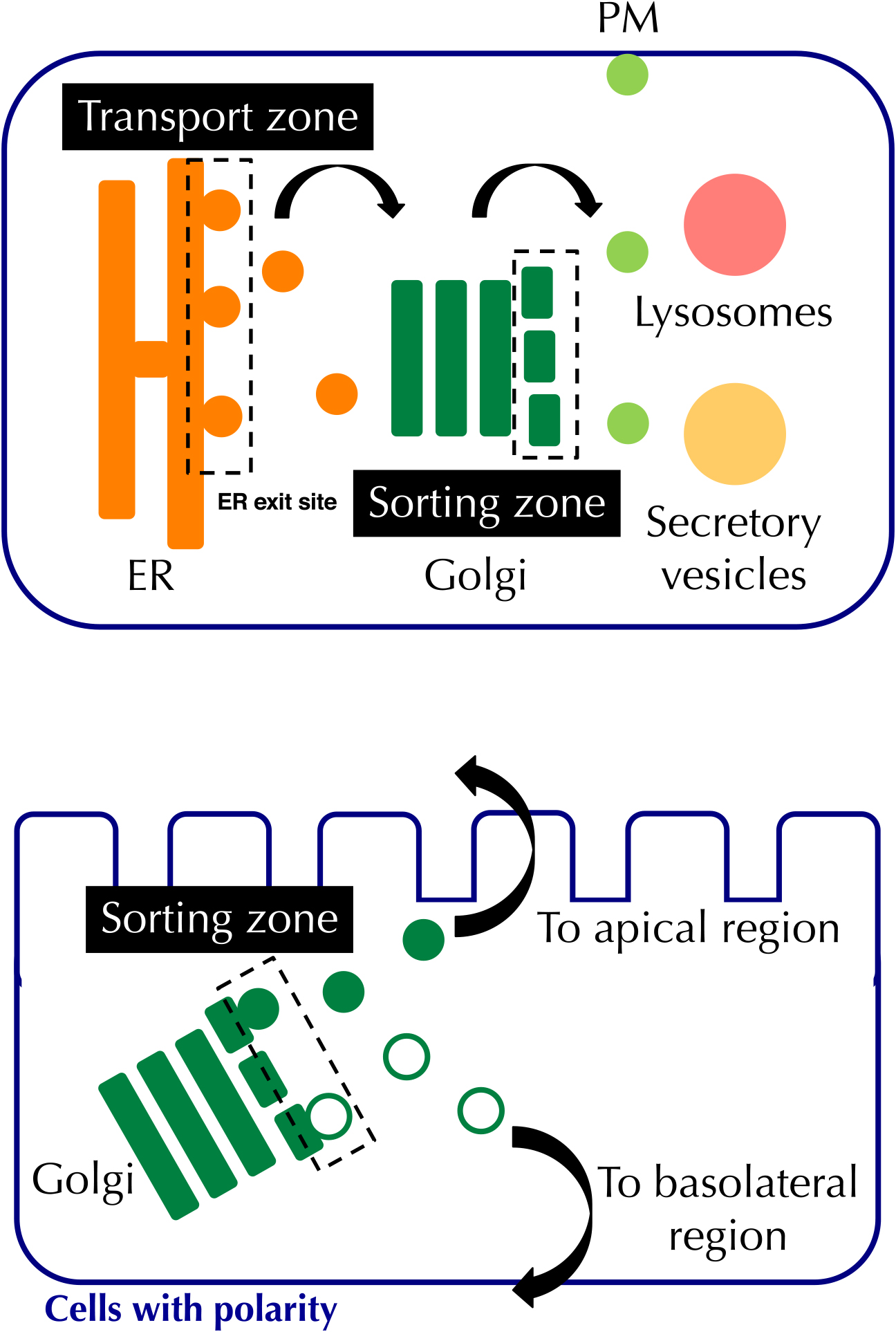
Organelle zones regulating vesicular transport in the ER and Golgi. Proteins synthesized in the ER are transported from the ER exit site to the Golgi, and sorted at the trans-Golgi and the trans-Golgi network to their final destinations.
In the ER, the ER exit site is a typical organelle zone for vesicular transport from the ER to the Golgi (Brandizzi and Barlowe, 2013; Sato and Nakano, 2005). Although the mechanism how the ER exit site is formed is unclear, it tends to form at the tip of the ER. In the Golgi, secretory and membrane proteins are sorted to their final destinations, such as the plasma membrane (PM), lysosomes, and secretory vesicles. In cells that have a polarity, proteins are sorted to apical and basolateral regions of the PM. Simons and colleagues reported that proteins are gradually sorted in the trans-Golgi and trans-Golgi network, and incorporated to distinct transport vesicles (Keller et al., 2001), suggesting the existence of a sorting zone in the Golgi. It is possible that the sorting zone closely cooperates with the folding zone as well as the post-modification zones including the GPI, proteoglycan and mucin zones described above.
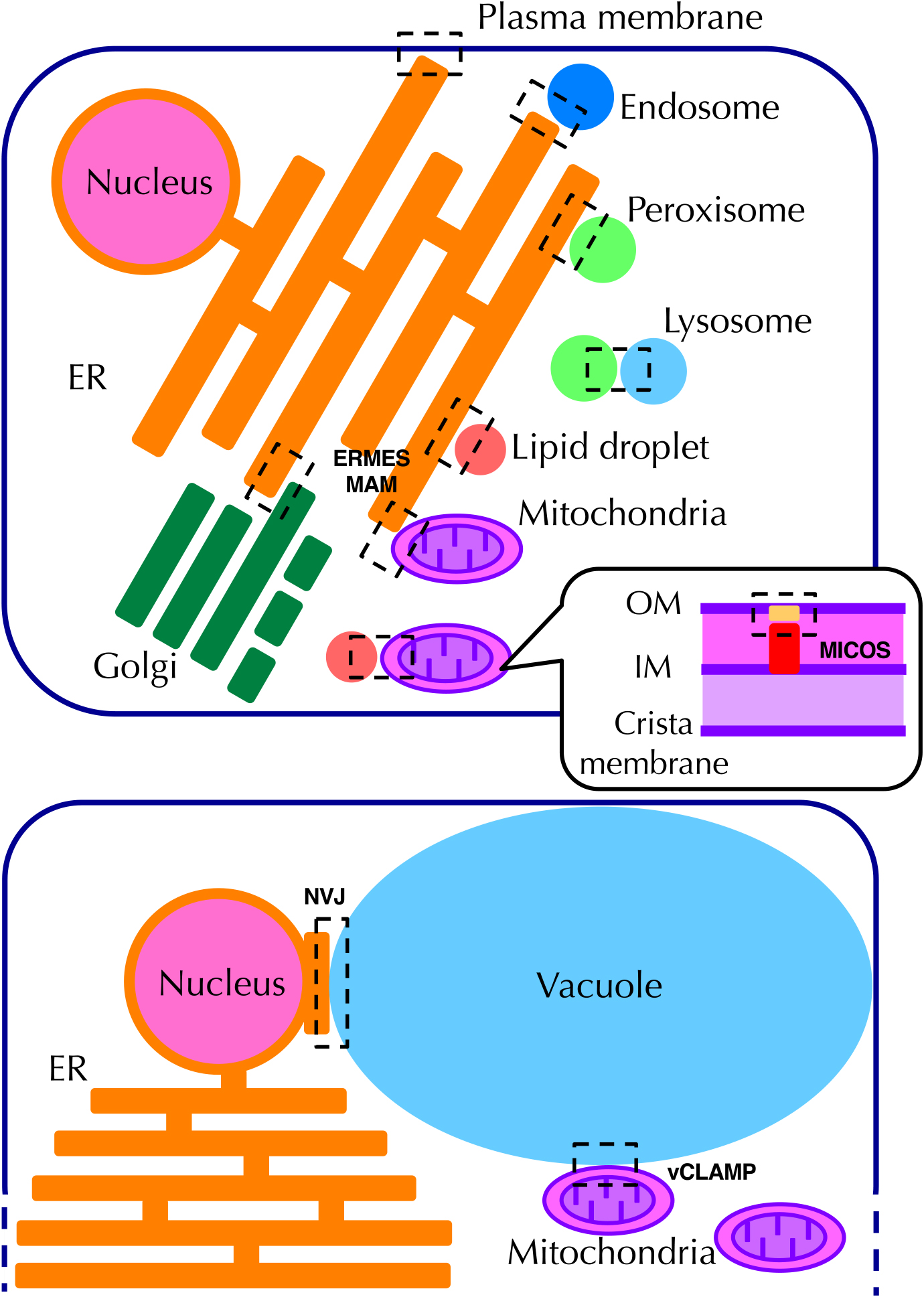
Organelle zones for organelle contact. Major organelle contact sites are indicated. Details are described in the text.
Physically interacting sites between organelles (organelle contact sites) are also a type of organelle zone. Various organelle contact sites have been reported (Eisenberg-Bord et al., 2016; Helle et al., 2013; Schrader et al., 2015), but we will describe only representative contact sites here.
(1) ER–PM contact siteThere are several reports on ER–PM contact sites. The first one was a contact site that transport lipids. Several oxysterol binding protein-related proteins (ORPs), such as Osh2 and Osh3, bind to ER vesicle-associated membrane protein-associated proteins (VAPs) and PM phosphoinositides through the FFAT and PH domains of ORPs, respectively. These ORPs transport membrane lipids from the ER to the PM and phosphatidylinositol-4-phosphate (PI4P) from the PM to the ER (Raychaudhuri and Prinz, 2010).
The second ER–PM contact site is mediated by STIM and Orai (Korzeniowski et al., 2009). STIM (an ER Ca2+ sensor) is an ER transmembrane protein involved in Ca2+ transport from the extracellular space to the ER lumen. In response to ER Ca2+ depletion, a C-terminal portion of STIM binds to PM phospholipids, resulting in the formation of a transient complex with Orai (a PM Ca2+ channel) and the influx of Ca2+ ions to the ER.
Tricalbins (Toulmay and Prinz, 2012) as well as Ist2 (Wolf et al., 2012) are also tethering factors at the ER–PM contact sites and are involved in the transport of glycerolipids and Ca2+ ions, but the precise mechanisms involved are as yet unclear.
(2) ER–mitochondria contact sitesThere are several types of contact sites between the ER and the mitochondria. The ER–mitochondria encounter structure (ERMES) complex transport membrane lipids in both directions using the SMP domains of its component proteins (Michel and Kornmann, 2012). Tethering factors of the ERMES complex include Mmm1, Mdm12, Mdm34, and Mdm10. Another ER–mitochondria contact site is mitochondria-associated ER membranes (MAM), which is mediated by the VDAC–GRP75–IP3R complex that transports Ca2+ ions from the ER to the mitochondria (Missiroli et al., 2018). Voltage dependent anion channel (VDAC) is a mitochondrial porin, whereas inositol trisphosphate receptor (IP3R) is an ER Ca2+ channel that opens upon inositol trisphosphate binding. GRP75 is a cytosolic chaperone that belongs to a HSP70 family, and tethers VDAC and IP3R. Moreover, mitofusin-2 (Koshiba et al., 2004), the VAPB–PTPIP51 complex (De Vos et al., 2012), and the Bap31–Fis1 complex (Iwasawa et al., 2011) separately form ER–mitochondrial contact sites, although their function remains controversial.
(3) Nucleus/ER–vacuole contact sitesThe nucleus–vacuole junction (NVJ) is a contact site between the nucleus and vacuole, and the NVJ is essential for piecemeal microautophagy of the nucleus (Kvam and Goldfarb, 2006). The tethering components of the NVJ are NVJ1 on the outer nuclear membrane and VAC8 on the vacuole membrane (Kvam and Goldfarb, 2006). At the NVJ, Osh1, which belongs to an ORP family, exchanges sterols for PI4P from the nucleus to the vacuole in a manner similar to other ORPs (Levine and Munro, 2001). NVJ2 is also localized at the NVJ and transports membrane lipids via its SMP domain (Toulmay and Prinz, 2011), though the precise mechanism is still unclear.
(4) ER–Golgi contact sitesCholesterol and ceramide are transported from the ER to the trans-Golgi. Oxysterol-binding protein (OSBP) binds both VAP proteins in the ER membrane and PI4P in the trans-Golgi membrane, and exchanges cholesterol for PI4P (Olkkonen, 2015). Ceramide transfer protein (CERT) exchanges ceramide for PI4P through a similar mechanism, by binding to ceramide, VAP proteins and PI4P through the START, FFAT, and PH domains (Yamaji and Hanada, 2014). OSBP and CERT are thought to be physically and functionally associated.
(5) Mitochondria–vacuole contact sitesVacuole and mitochondria patch (vCLAMP) transports lipids between the mitochondria and other endomembrane system (Elbaz-Alon et al., 2014). Tethering factors of vCLAMP include Vps39, Ypt7, and Vps13, and the complex is enriched with ion and amino acid transporters.
(6) Mitochondrial OM–IM contact sitesMost mitochondrial proteins are synthesized in the cytosol and transported to the mitochondrial matrix through the mitochondrial outer membrane (OM) and inner membrane (IM) contact sites (Endo et al., 2011). Its tethering factors comprise the mitochondrial contact site and cristae organizing system (MICOS) in the IM and MICOS interaction partners in the OM (Kozjak-Pavlovic, 2017).
(7) Other contact sitesPeroxisomes are the organelle where long chain fatty acids are degraded, and some peroxisomal membrane proteins are synthesized in the ER and then transported to the peroxisomes (Fujiki, 2016). Pex3 and Inp1 form the ER–peroxisome contact site, which is essential for peroxisome inheritance (Munck et al., 2009). The ER–endosome contact site is formed by tethering factors such as VAP proteins, ORP1L, and ORP5, which is similar to OSBP at the ER–Golgi contact site (Du et al., 2011). This contact site may be important for the transport of membrane lipids. Synaptotagmin-7 mediates tethering at the lysosome–peroxisome contact site, which transports cholesterol from the lysosomes to the peroxisomes (Chu et al., 2015). ER–lipid droplet and mitochondria–lipid droplet contact sites require DGAT2 and Perilipin-5, respectively (Xu et al., 2012). The former contact site is essential for the import of lipids from the ER to lipid droplets.
Apoptosis is an indispensable mechanism for multicellular organisms (Miura, 2012). During the process of apoptosis, cells release pro-apoptotic factors such as cytochrome c, Omi, Smac/DIABLO, AIF, HtrA2, and EndoG from the mitochondria into the cytosol (Nagata and Tanaka, 2017). To release these factors, the permeability of the mitochondrial membrane should be increased, which is regulated by two Bcl-2 family proteins, BAK and BAX. In normal situations, BAK exists throughout the mitochondrial OM, whereas BAK forms clusters in which the diameter is around 20 nm (Nasu et al., 2016). These BAK clusters are called the apoptosis zone (Shimizu, 2019), although it is uncertain whether the cluster itself is a channel for cytochrome c release. Interactions between BAK and other Bcl-2 family proteins as well as VDAC in the OM and cardiolipins in the IM are essential for formation of the apoptosis zone.
OASIS is a transcription factor that belongs to the ATF6 family (Murakami et al., 2009). In normal conditions, OASIS resides in the ER membrane, whereas OASIS is localized in blebs of the nuclear envelope in response to DNA damage stress, which is termed the nucleus–ER cooperating zone (Dr. Kazunori Imaizumi and colleagues, personal communication). Dr. Tatsuya Saitoh and colleagues identified the innate immune zone in the Golgi apparatus, where STING accumulates and undergoes palmitoylation to regulate the innate immune response upon invasion of pathogens (Takahama et al., 2017).
Organelles are not static, rather they are very dynamic. For example, the ER remarkably expands during the differentiation of plasma cells, in order to support the synthesis of a number of immunoglobulin molecules. The mechanism that upregulates the capacity of organelles is collectively called “organelle autoregulation” (Sasaki and Yoshida, 2015). The ER stress response (Kimata and Kohno, 2011; Mori, 2015), Golgi stress response (Taniguchi and Yoshida, 2017), and lysosome stress response (Napolitano and Ballabio, 2016) constitute organelle autoregulation of the ER, Golgi, and lysosome, respectively. Because each organelle consists of a set of organelle zones, it is highly possible that the capacity of each organelle zone is controlled by distinct regulatory mechanisms. For instance, it is possible that the folding zone in the ER is upregulated when folding capacity becomes insufficient, whereas the ERAD zone is augmented if ERAD capacity becomes insufficient. Actually, the expression of ER chaperones is regulated by the ATF6 pathway of the ER stress response, whereas that of ERAD factors is mainly regulated by the IRE1 pathway (Yamamoto et al., 2007). Recently, Yoshida and colleagues revealed that the proteoglycan pathway of the Golgi stress response regulates the capacity of the proteoglycan zone (Sasaki et al., 2019). Because autoregulation of the capacity for each organelle is essential for cells to orchestrate their function in accordance with various environmental situations, research on organelle zone autoregulation will become an increasingly important issue in cell biology.
We thank Ms. Mikiko Ochiai for secretarial assistance. This work was supported by JSPS KAKENHI (Grant numbers 19K06645, 19K16131 and JP17J00067, a Grant-in-Aid for Scientific Research on Innovative Areas of MEXT (JP17H06414) and JSPS Bilateral Joint Research Projects.