Abstract
The T-box 19 (TBX19) gene encodes a transcription factor characterized by a highly conserved DNA-binding motif (T-box). Recent studies have revealed that TBX19 has been identified as one of the genes activated by KRAS mutations, and is upregulated in colon adenoma. These results indicate that TBX19 may work as an oncogene in colorectal cancer (CRC). However, the expression and role of TBX19 have yet to be investigated. Here, we investigated TBX19 mRNA and protein expressions in colon cancer cells or surgically resected CRC. We found that TBX19 mRNA expression was significantly increased in tumorous tissues compared to that in non-tumorous tissues, and increased TBX19 mRNA expression was associated with positive lymph node metastasis in our cohort. The expression of TBX19 mRNA was not correlated with that of TBX19 protein in tissue sample taken from the CRC patients. Moreover, TBX19 showed positive staining even in the normal colonic tissues and the adjacent non-tumorous tissues. These results suggest that the expression of TBX19 protein is not correlated with the expression of TBX19 mRNA. In addition, our results promote further investigations into the impact of TBX19 upregulation on colorectal carcinogenesis, as well as the underlying mechanisms.
Introduction
Colorectal cancer (CRC) is the third most common type of cancer that occurs in both men and women all over the world1,2). One of the most important molecular pathogeneses of colorectal carcinogenesis is the adenoma-carcinoma sequence1). The adenoma-carcinoma sequence is stepwise genetic aberrations, including APC, KRAS and TP53 mutations, in CRC patients3). APC mutations are an early event in this multistep process, followed by KRAS-activating mutations and TP53-inactivating mutations3). While chemotherapy and molecular targeting drugs for CRC treatment have progressed recently, genetic aberrations among APC, KRAS, and TP53 are not therapeutic targets4,5). It is particularly necessary to develop therapeutic agents for KRAS mutations as they are frequently detected in CRC patients, as well as other malignant tumors, to improve cancer mortality6). Although there have been several studies on the development of targeted drugs for KRAS mutations7-9), they have not yet been used in a clinical setting.
The T-box (TBX) gene family encodes a large family of transcription factors and plays a fundamental role in early embryogenesis during the developmental process10). Several studies have revealed aberrations of TBX genes in inherited human disorders, such as TBX1 mutation in DiGeorge syndrome, TBX3 mutation in Ulnar-Mammary syndrome, TBX5 mutation in Holt-Oram syndrome, and TBX22 mutation in cleft palate with ankyloglossia11). In addition, recent studies have also found that TBX genes may be associated with cancer development in various malignant tumors12). TBX2 and TBX3, which are downstream targets of the Wnt/beta-catenin pathway, are frequently mutated in ovarian cancer and are amplified in breast cancer. Furthermore, they are associated with hepatocellular carcinoma, pancreatic cancer, and malignant melanoma13-18).
In the present study, we focused on the analysis of TBX19, which is expressed in the rostral ventral diencephalon and pituitary gland19). TBX19 mutations lead to a lack of adrenocorticotrophin resulting in adrenal insufficiency20). On the other hand, TBX19 has been identified as one of the genes activated by KRAS mutation, and is upregulated in colon adenoma21,22). These results indicate that TBX19 might work as an oncogene in CRC, but the expression and role of TBX19 in CRC remain unknown. Here, we investigated TBX19 mRNA and protein expressions in surgically resected CRC tissues, and examined the biological significance.
Materials and Methods
Clinical samples of patients
A total of 89 surgical specimens obtained from CRC patients who had undergone surgical resection at Fukushima Medical University Hospital between January 2008 and December 2010 were used for the experiments. All 89 cases are used for comprehensive gene expression analysis, 5 cases are used for protein expression analysis by western blotting, and 54 cases are used for immunohistochemical (IHC) staining. In addition, 3 cases of adenoma were used for IHC staining. Information regarding age, sex, TNM stage, and pathological diagnosis, including lymphatic and venous invasion, were retrospectively collected. The carcinomas at the time of primary tumor resection were staged according to the Union for International Cancer Control UICC classification (the 7th classification)23,24). Written informed consent was obtained from all patients. This study was approved by the ethics committee of Fukushima Medical University.
Comprehensive gene expression analysis
TBX19 expression data were obtained using custom microarray analysis as previously described25,26). In brief, the surgical specimen was homogenized and mixed with ISOGEN reagent (NIPPON GENE, Tokyo, Japan). Total RNA was subjected to purification of polyA(A)+RNA using MicroPoly(A) Purist Kit (Thermo Fisher Scientific, Waltham, MA, USA). The human common reference RNA was prepared by mixing equal amounts of poly(A)+ RNA extracted from 22 human cancer cell lines (A431, A549, AKI, HBL-100, HeLa, HepG2, HL60, IMR-32, Jurkat, K562, KP4, MKN7, NK-92, Raji, RD, Saos-2, SK-N-MC, SW-13, T24, U251, U937, and Y79).
Synthetic polynucleotides (80-mers) representing 31,797 human transcripts (MicroDiagnostic, Tokyo, Japan) were arrayed on aminosilane-coated glass slides with a custom-made arrayer. RNA (2 μg) was subjected to reverse transcription with SuperScript II (Thermo Fisher Scientific). Sample RNA was labeled using Cyanine 5-dUTP (Perkin-Elmer, Boston, MA, USA) and reference RNA was labeled using Cyanine 3-dUTP. Hybridization was performed with a labeling and hybridization kit (MicroDiagnostic). Signals were measured with a GenePix 4000B scanner (Axon Instruments, Union City, CA, USA) and then processed into primary expression ratios. The primary expression ratios were then converted into log2 values and compiled into a matrix. We assigned an expression ratio of 1 (log ratio of 0) for spots that exhibited fluorescence intensities under the detection limits, and we included these in the signal calculation of the mean averages. Data were processed by MDI gene expression analysis software package (MicroDiagnostic).
Cell line culture
The colon cancer cell lines used in this study were originally obtained from the American Type Culture Collection (Rockville, MD, USA) and were cultured in the recommended media with 10% fetal bovine serum. These monolayer cells were maintained in a 37°C incubator with 5% CO2. Cells were checked regularly under a light microscope and subcultured once they had reached 80% to 90% confluence.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions as previously described27). Complementary DNA (cDNA) was synthesized from 5 μg of total RNA with a random hexamer using the SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific). These cDNAs were used for the measurement of gene expression with a 7500 Real-time PCR system (Thermo Fisher Scientific) using TaqMan probes. The assessors were blinded to patient information and performed experiments in triplicate. Taqman expression assays were purchased from Thermo Fisher Scientific;TBX19 (Hs01113611_m1) and β-actin (Hs99999903_m1). β-actin was used as an internal control. Relative TBX19 gene expression was calculated using the 2-ΔΔCT method, according to the supplier’s protocol (Thermo Fisher Scientific)28).
Western blotting
Cancer cell lines and surgical specimens were homogenized in a 100 mM Tris-HCl (pH 7.6) buffer containing 0.15 M NaCl, 5 mM EDTA, 1% Triton X-100, 5% glycerol by Polytron PT3100 homogenizer (Kinematica AG, Luzern, Switzerland). After centrifugation at 17,400 xg for 15 min at 4°C, the supernatants were collected. Then, 20 ug of each protein sample was run on SDS-polyacrylamide gels (5-15% gradient;Thermo Fisher Scientific) and blotted onto Immun-Blot PVDF membranes (Bio-Rad Laboratories, Hercules, CA, USA). The blotted membranes were incubated with the indicated primary antibodies overnight at 4°C. Rabbit monoclonal anti-TBX19 (HPA005800, Sigma-Aldrich, St. Louis, MO, USA) was used at 1: 500 dilution, while mouse monoclonal anti-β-actin antibody (sc-69879, Santa Cruz Biotechnology, USA) was used as a loading control at 1: 2,500 dilution. The blotted membranes were then incubated with the appropriate horseradish peroxidase (HRP)-conjugated goat-anti-mouse IgG (sc-2005, Santa Cruz Biotechnology) secondary antibody at a dilution of 1: 5,000. Signals were detected by ImageQuant LAS4000 (GE Healthcare Bio-sciences, Pittsburgh, PA, USA) using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific).
Immunohistochemical staining and evaluation
IHC staining was carried out on paraffin-embedded histological sections (4 μm thick) using a polymer peroxidase method. Briefly, after deparaffinization and rehydration, the sections were treated with 0.3% hydrogen peroxide in methanol for 30 min to block endogenous peroxidase activity. Following rinsing in phosphate-buffered saline (PBS) (Thermo Fisher Scientific), the sections were incubated with anti-TBX19 antibody (HPA005800, 1: 2,000 dilution; Sigma-Aldrich) at 4°C overnight. Three further washes (5 min per wash) in PBS was followed by treatment with a peroxidase-labeled polymer, conjugated to goat anti-rabbit immunoglobulins (Dako EnVision+System-HRP Labelled Polymer; ready-to-use;#K4003;Dako;Agilent Technologies), as the secondary antibody for 30 min at room temperature. The staining was visualized with diaminobenzidine, followed by counterstaining with hematoxylin. Expression of these proteins was evaluated as positive when the nucleus of tumorous tissues and the total field of view were observed at 400×magnification. We evaluated the staining of each specimen. The rate of positively stained cells was counted among three randomly selected fields (200 μm×200 μm) in the tumorous and non-tumorous tissues. Cytoplasmic granular signals were counted in the tumorous tissues.
Statistical Analysis
Data were presented as the mean±SD. Statistical analyses were performed using GraphPad Prism 6 software (GraphPad Software, La Jolla, CA, USA). The Mann-Whitney U test and Wilcoxon matched pairs test were used for comparison of the means of the two groups and Kruskal-Wallis test was used for comparison of more than two groups. Log-rank test was used for survival comparisons. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression analysis of TBX19 from the comprehensive analysis data
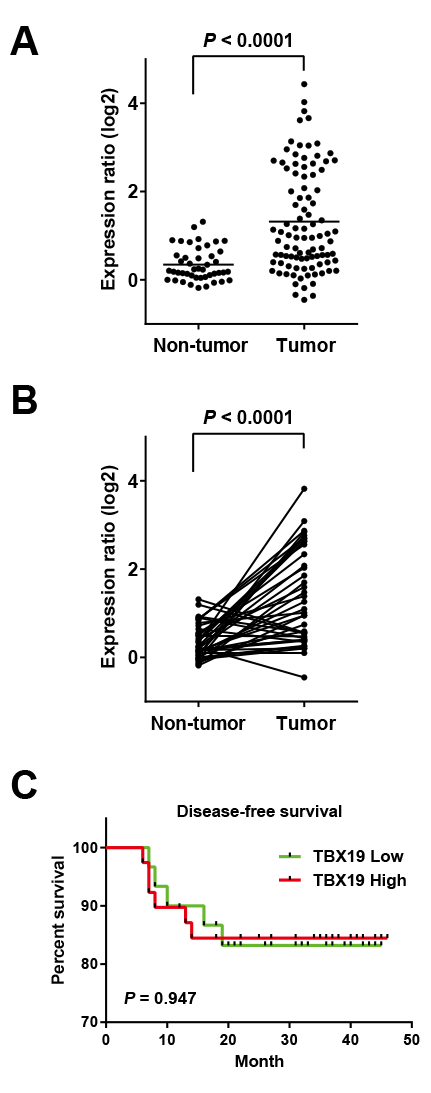
Firstly, the mRNA level of TBX19 in the CRC specimen was determined by using comprehensive gene expression analysis data. The expression ratios of TBX19 were compared between 89 tumorous tissue samples and 60 non-tumorous tissue samples, which revealed that TBX19 had a significantly higher expression in tumorous tissues compared to non-tumorous tissues (P<0.0001, Mann-Whitney’s U-test) (Fig. 1A). Of note, among available 40 pairs of non-tumorous and tumorous tissues, upregulation of TBX19 was also observed in the tumorous tissues compared to the non-tumorous tissues (P<0.0001, Wilcoxon matched pairs test) (Fig. 1B).
Next, we analyzed TBX19 expression levels with clinicopathological factors in the CRC specimens (Table 1). The case with positive lymph node metastasis showed significantly higher expression of TBX19 (P=0.012). However, TBX19 expression level was not found to be associated with age, gender, TNM stage, histology, tumor depth, lymphatic invasion, venous invasion, or distant metastasis. Kaplan-Meier analysis demonstrated no association between increased TBX19 levels and relapse-free survival (P=0.9473, log-rank test) (Fig. 1C).
TBX19 mRNA and protein expression
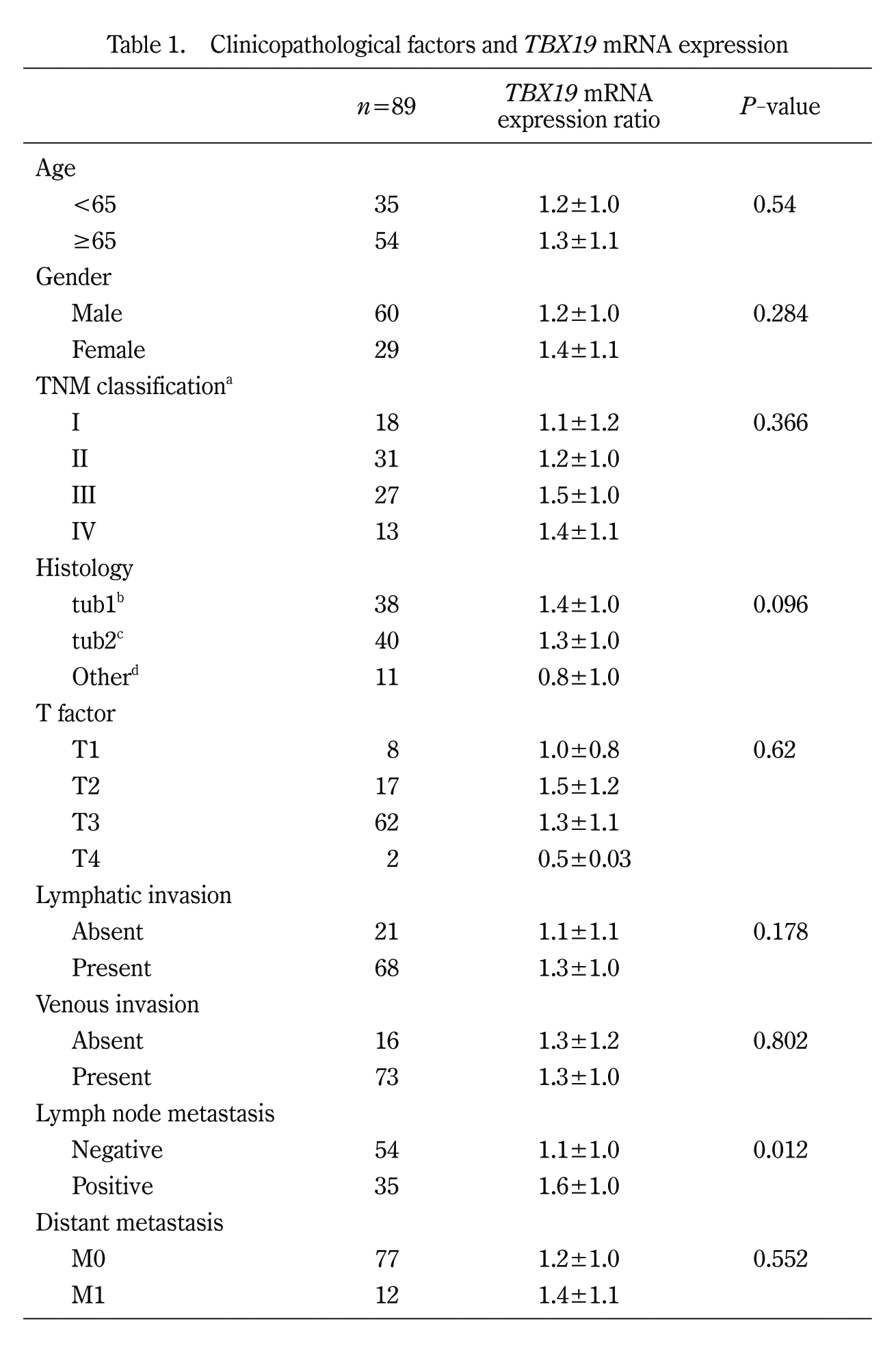
To further confirm that TBX19 mRNA expression is upregulated in CRC, we investigated TBX19 expression in 7 colon cancer cell lines. TBX19 mRNA expression was investigated by real-time PCR, and TBX19 protein expression was investigated by western blotting for the 7 colon cancer cells (Fig. 2A). Consistent with TBX19 mRNA expression, TBX19 protein expression was upregulated in the LS180, LS174T, and SW837 cells. Then, we analyzed TBX19 protein expression by western blotting in three representative non-tumorous/tumorous CRC tissues that showed high TBX19 mRNA expression (Fig. 2B). However, TBX19 protein was not highly expressed in the tumorous tissues compared to the non-tumorous tissues. These results indicate that the upregulation of TBX19 mRNA did not result in upregulation of TBX19 protein expression.
IHC staining for TBX19
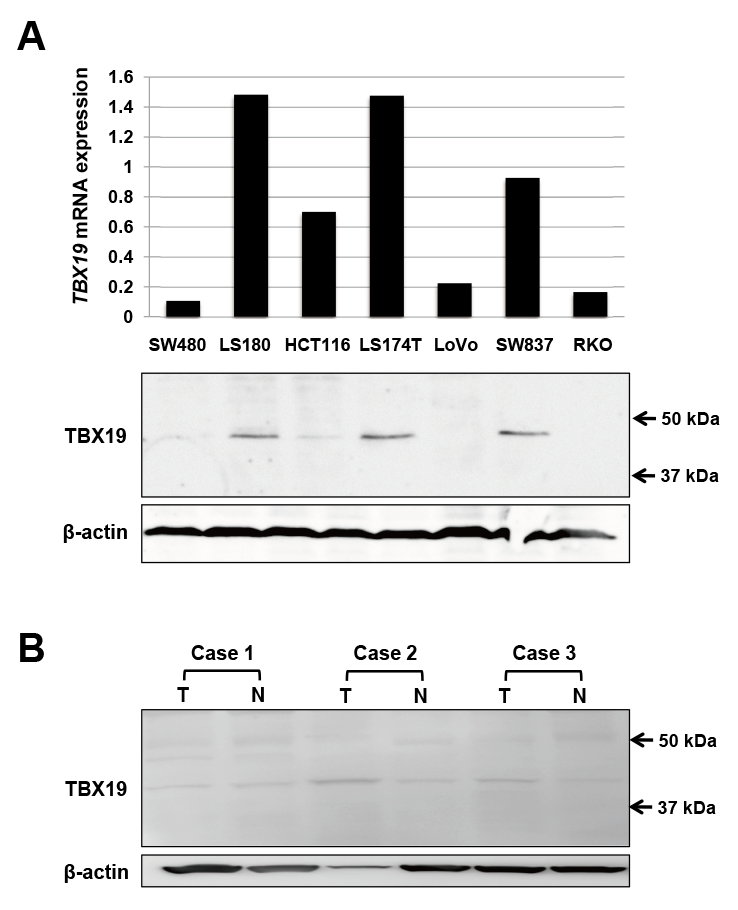
Next, we performed IHC staining for TBX19 in 3 adenoma and 54 CRC specimens. While normal colonic mucosa exhibited weak TBX19 staining (Fig. 3A), all 3 adenoma showed positive TBX19 staining (Fig. 3B). In the CRC specimens, TBX19 expression was observed in the nucleus of tumorous and adjacent non-tumorous cells (Fig. 3C). When we assessed TBX19 staining intensity, the tumorous tissue samples showed a higher percentage of positive TBX19 cells compared to the non-tumorous tissue samples (Fig. 3D). However, this staining intensity was not associated with any clinicopathological factors in our cohort (Table 2). We additionally performed TBX19 staining for metastatic liver cancer from CRC. The metastatic liver tumor showed positive staining for TBX19, but normal liver tissue showed negative TBX19 staining (Fig. 3E). These results suggested that TBX19 was specifically upregulated in CRC cells.
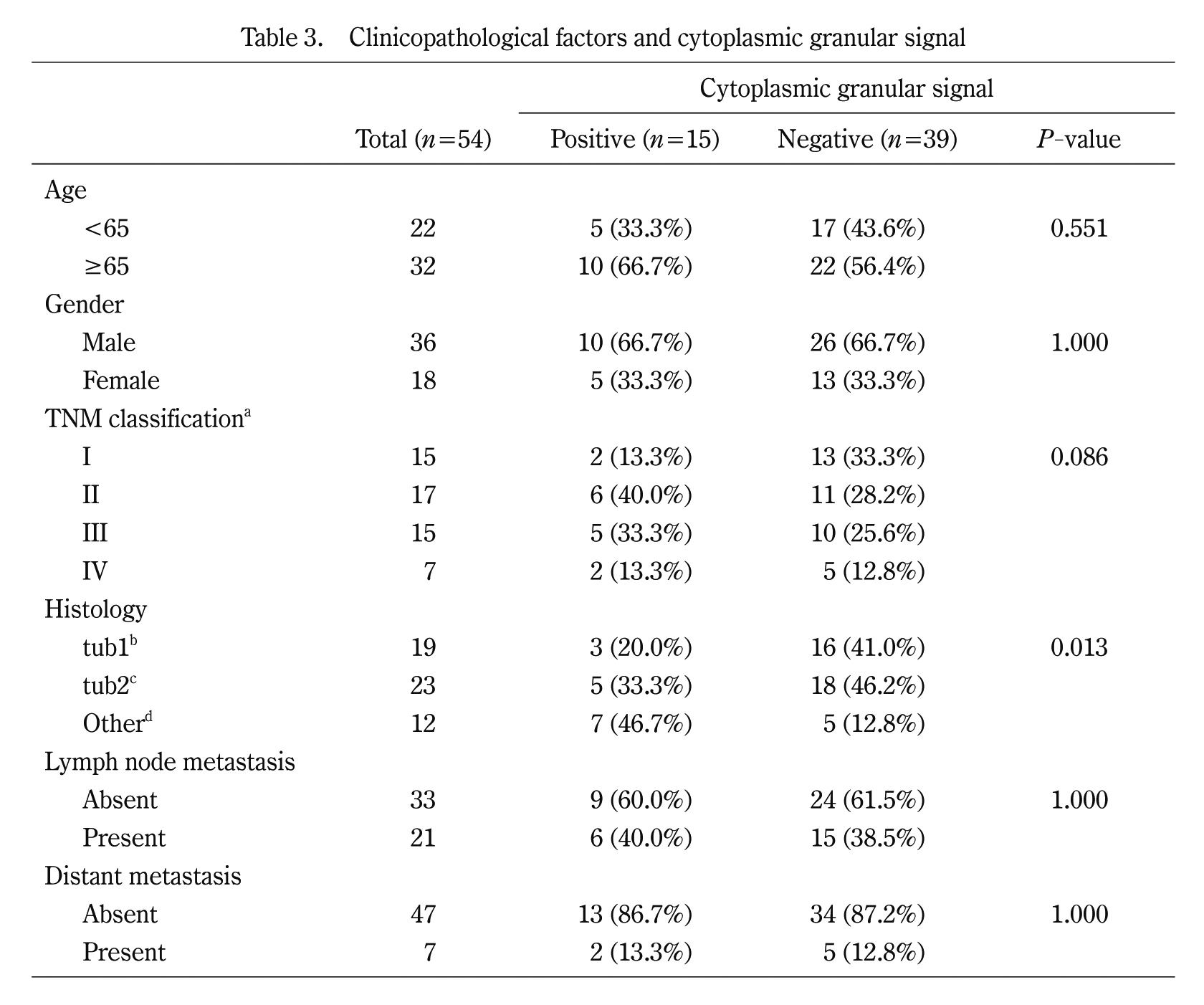
To further understand TBX19 staining, we focused on the cytoplasmic granular signals in CRC cells (Fig. 3F). While the cytoplasmic granular signals in cytoplasmic tumor cells were not observed in the normal tissue, they were observed in 15 of the 54 (27.8%) CRC tissues. The signals tended to be observed in undifferentiated histological types, such as poorly differentiated or mucinous adenocarcinoma (Table 3). These results further suggest that TBX19 mRNA was upregulated in CRC and associated with worse CRC outcomes.
Discussion
In the present study, we found that TBX19 mRNA was upregulated in CRC and increased expression of TBX19 mRNA was associated with positive lymph node metastasis in CRC patients. On the contrary, TBX19 protein expression was not upregulated in CRC and was not associated with any clinicopathological factors in our cohort. Therefore, our study shows that, while upregulated TBX19 mRNA may have a pivotal role in colon tumorigenesis, the role of TBX19 protein in colonic tumorigenesis is still unknown and need a further consideration. Furthermore, our results also suggest that the expression of TBX19 protein was not correlated with TBX19 mRNA expression. This was further confirmed by the result that the CRC patients, which showed high TBX19 mRNA expression, did not exhibit higher TBX19 protein expression in the tumorous tissues than in the non-tumorous tissues by Western blotting in our small number of cases. Even in the normal colonic tissues and the adjacent non-tumorous tissues showed weak positive staining for TBX19, suggesting that TBX19 protein may have a role in keeping normal mucosal homeostasis and in affecting CRC tumor development. Therefore, the evaluation of TBX19 in CRC by IHC staining requires further investigation. Of course, the evaluation of sensitivity and specificity for anti-TBX19 antibody remains to be required. When observing IHC staining, we were interested in the cytoplasmic granular signals in the tumor cells and found that they tended to associate with tumor differentiation in our cohort. To date, because the mechanism for the formation of granular signals in tumor cells has not yet been fully understood, further morphological and functional studies are required.
Consistent with previous reports, TBX19 was also upregulated in adenoma21,22). Because adenoma is considered to be a type of pre-cancerous tumor, we believe that TBX19 plays a role in the adenoma-carcinoma sequence. A previous in vitro experiment revealed that TBX19 is one of the downstream genes that are activated by KRAS mutations21). In fact, TBX19 was highly expressed in most colorectal adenomas with activated KRAS mutations22). KRAS mutations are one of the steps in the adenoma-carcinoma sequence that accumulate several genetic aberrations in CRC. Therefore, because KRAS mutations occur in both colorectal adenoma and cancer, it is understandable that TBX19 expression is upregulated in CRC as well as in adenoma cases. In CRC treatment, the mutational status of KRAS is only used as a predictive marker for the effectiveness of anti-EGFR antibodies, cetuximab, or panitumumab29). This is due to KRAS being a downstream effector of EGFR. In addition, and most importantly, KRAS mutations have not yet to be a direct druggable target. KRAS mutations are the most frequently occurring aberrations in human cancer, including CRC6,30). In spite of huge efforts to develop molecular targeting drugs for KRAS mutations7), no useful agents have yet been included in clinical treatment strategies for CRC. With recent progress in the understanding of KRAS biology, KRAS has been considered as druggable by either targeting the mutations directly or targeting the downstream pathways, such as RAF-MAPK and PIK3K31). Of note, because TBX19 is also downstream of KRAS, further experimental or mice studies investigating the functional role of TBX19 may provide a therapeutic opportunity for KRAS-driven cancers.
In conclusion, the current study reports the TBX19 expression status in CRC. Our results suggest that TBX19 as a candidate therapeutic target for CRC.
Acknowledgements
This research was partially supported by grants for translational research programs from New Energy and Industrial Technology Development Organization (NEDO) (Tokyo, Japan).
Conflict of Interest
The authors declare that there is no conflict of interest.
References
- 1. Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet, 383: 1490-1502, 2014.
- 2. Hori M, Matsuda T, Shibata A, et al. Cancer incidence and incidence rates in Japan in 2009:a study of 32 population-based cancer registries for the Monitoring of Cancer Incidence in Japan (MCIJ) project. Jpn J Clin Oncol, 45: 884-891, 2015.
- 3. Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature, 386: 623-627, 1997.
- 4. Tournigand C, Andre T, Achille E, et al. FOLFIRI followed by FOLFOX6 or the reverse sequence in advanced colorectal cancer:a randomized GERCOR study. J Clin Oncol, 22: 229-237, 2004.
- 5. Goldberg RM, Sargent DJ, Morton RF, et al. A randomized controlled trial of fluorouracil plus leucovorin, irinotecan, and oxaliplatin combinations in patients with previously untreated metastatic colorectal cancer. J Clin Oncol, 22: 23-30, 2004.
- 6. Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature, 502: 333-339, 2013.
- 7. Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature, 503: 548-551, 2013.
- 8. Zimmermann G, Papke B, Ismail S, et al. Small molecule inhibition of the KRAS-PDEdelta interaction impairs oncogenic KRAS signalling. Nature, 497: 638-642, 2013.
- 9. Dietlein F, Kalb B, Jokic M, et al. A Synergistic Interaction between Chk1- and MK2 Inhibitors in KRAS-Mutant Cancer. Cell, 162: 146-159, 2015.
- 10. Papaioannou VE. The T-box gene family: emerging roles in development, stem cells and cancer. Development, 141: 3819-3833, 2014.
- 11. Packham EA, Brook JD. T-box genes in human disorders. Hum Mol Genet, 12 Spec No 1: R37-44, 2003.
- 12. Wansleben S, Peres J, Hare S, Goding CR, Prince S. T-box transcription factors in cancer biology. Biochim Biophys Acta, 1846: 380-391, 2014.
- 13. Rowley M, Grothey E, Couch FJ. The role of Tbx2 and Tbx3 in mammary development and tumorigenesis. J Mammary Gland Biol Neoplasia, 9: 109-118, 2004.
- 14. Lu J, Li XP, Dong Q, Kung HF, He ML. TBX2 and TBX3:the special value for anticancer drug targets. Biochim Biophys Acta, 1806: 268-274, 2010.
- 15. Abrahams A, Parker MI, Prince S. The T-box transcription factor Tbx2:its role in development and possible implication in cancer. IUBMB Life, 62: 92-102, 2010.
- 16. Douglas NC, Papaioannou VE. The T-box transcription factors TBX2 and TBX3 in mammary gland development and breast cancer. J Mammary Gland Biol Neoplasia, 18: 143-147, 2013.
- 17. Hansel DE, Rahman A, House M, et al. Met proto-oncogene and insulin-like growth factor binding protein 3 overexpression correlates with metastatic ability in well-differentiated pancreatic endocrine neoplasms. Clin Cancer Res, 10: 6152-6158, 2004.
- 18. Vance KW, Carreira S, Brosch G, Goding CR. Tbx2 is overexpressed and plays an important role in maintaining proliferation and suppression of senescence in melanomas. Cancer Res, 65: 2260-2268, 2005.
- 19. Liu J, Lin C, Gleiberman A, et al. Tbx19, a tissue-selective regulator of POMC gene expression. Proc Natl Acad Sci U S A, 98: 8674-8679, 2001.
- 20. Yi CH, Terrett JA, Li QY, et al. Identification, mapping, and phylogenomic analysis of four new human members of the T-box gene family: EOMES, TBX6, TBX18, and TBX19. Genomics, 55: 10-20, 1999.
- 21. Chen YF, Wang JY, Wu CH, Chen FM, Cheng TL, Lin SR. Detection of circulating cancer cells with K-ras oncogene using membrane array. Cancer Lett, 229: 115-122, 2005.
- 22. Wang JY, Wang YH, Jao SW, et al. Molecular mechanisms underlying the tumorigenesis of colorectal adenomas:correlation to activated K-ras oncogene. Oncol Rep, 16: 1245-1252, 2006.
- 23. Sobin LH, Compton CC. TNM seventh edition: what’s new, what’s changed:communication from the International Union Against Cancer and the American Joint Committee on Cancer. Cancer, 116: 5336-5339, 2010.
- 24. Sobin LH GM, Wittekind Ch. International Union Against Cancer (UICC) TNM Classification of Malignant Tumors. 7th ed. Oxford, UK:Wiley-Blackwell, 2009.
- 25. Okabe N, Ezaki J, Yamaura T, et al. FAM83B is a novel biomarker for diagnosis and prognosis of lung squamous cell carcinoma. Int J Oncol, 46: 999-1006, 2015.
- 26. Tachibana K, Saito M, Imai JI, et al. Clinicopathological examination of dipeptidase 1 expression in colorectal cancer. Biomed Rep, 6: 423-428, 2017.
- 27. Saito M, Shiraishi K, Matsumoto K, et al. A three-microRNA signature predicts responses to platinum-based doublet chemotherapy in patients with lung adenocarcinoma. Clin Cancer Res, 20: 4784-4793, 2014.
- 28. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 25: 402-408, 2001.
- 29. Siddiqui AD, Piperdi B. KRAS mutation in colon cancer:a marker of resistance to EGFR-I therapy. Ann Surg Oncol, 17: 1168-1176, 2010.
- 30. Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature, 487: 330-337, 2012.
- 31. McCormick F. KRAS as a Therapeutic Target. Clin Cancer Res, 21: 1797-1801, 2015.