2018 年 6 巻 1 号 p. 44-50
2018 年 6 巻 1 号 p. 44-50
In this study, a collection of Salmonella enterica subspecies obtained from live mice caught on 32 poultry farms in the Northeast US between 1995 to 1998 was evaluated to provide a historical reference for serotype distribution during a time when egg contamination by serotype Enteritidis was at its peak. Of 821 mice cultured, 157 were positive (19.1%). Seven mice harbored two serotypes of Salmonella. Nine serotypes were detected, eight of which are often associated with foodborne illness. The three most prevalent serotypes were Enteritidis, Heidelberg, and Typhimurium. Enteritidis and Typhimurium were obtained from both spleens and intestines without preference according to type of sample. In contrast, Heidelberg was isolated most often from intestines and Schwarzengrund was most often obtained from spleens. These results support that the house mouse Mus musculus was a risk factor for introduction of multiple pathogenic Salmonella serotypes in poultry raised in the Northeast US during the mid-1990s. Isolates were submitted to the Food and Drug Administration and draft genomes for 64 isolates of Salmonella enterica serovar Enteritidis data have been released through the National Center for Biotechnology Information via the GenomeTrakr network.
Whole Genome Sequencing (WGS) is useful for tracking sources of foodborne outbreaks when it is combined with geographic metadata and epidemiological investigations1,2). WGS databases are improved by including historical and current collections of isolates, because evolutionary trends are impacted by time and local conditions. Including information from different time periods and regions to assess trends improves accuracy. Regulatory agencies in the US charged with evaluating foodborne risk of illness have set a goal of completing a million genomes of Salmonella enterica by 2020. The quality of databases matter, and a continuing challenge is to include information that provides a broad range of information without letting databases become overly skewed towards a single serotype or industry or host3).
Salmonella enterica subspecies I is the causative agent of foodborne human salmonellosis, but only 30 of over 1500 serotypes are frequently implicated in outbreaks4,5,6). Serotype Enteritidis is the world’s leading cause of foodborne illness, and it is tracked closely in epidemiological investigations along with other major serotypes such as serotypes Typhimurium, Heidelberg, and Newport. Enteritidis is especially problematic because it migrates through the tissues of the hen, colonizes reproductive tract organs, and survives in the internal contents of eggs7,8,9). The large outbreaks caused by serotype Enteritidis in association with shell eggs between 1980 and 2000 resulted in implementation of stringent biocontrol programs on poultry farms, and these measures included rodent control10). Mice are ubiquitous inhabitants of farms, urban areas and homes, and they are a challenging vector of foodborne disease to control6,11). Despite implementation of stringent control programs, reductions in outbreaks, from Salmonella enterica in general and serotype Enteritidis in particular, per capita reduction in foodborne salmonellosis appears to have reached a plateau in the United States around 2004 and in Europe around 2009 (Fig. 1)12). Thus, continued reduction of foodborne illness caused by Salmonella enterica requires persistent, thorough and novel application of efforts if continued reduction is to be achieved.
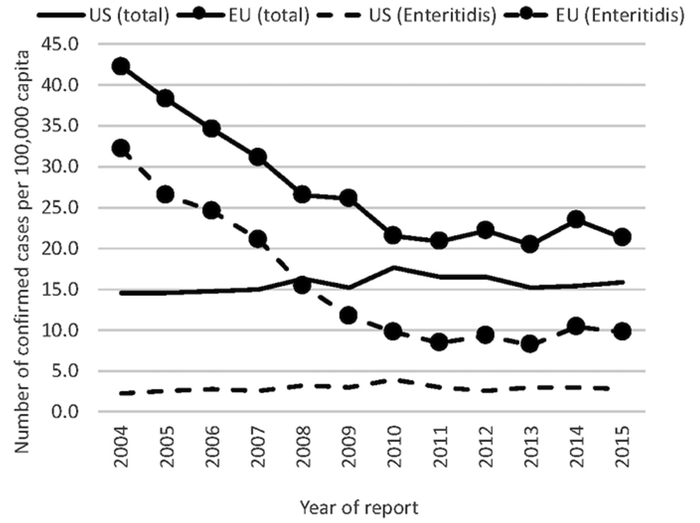
Collated graph of the reported case incidence per 100,000 capita in the United States and Europe for total Salmonella enterica and serotype Enteritidis
Solid lines, total Salmonella all serotypes; hatched lines, Salmonella serotype Enteritidis only. Black lines represent US data, black lines with circle indicate EU data. For the US, data were obtained from the Centers for Disease Control FoodNet sites for the year indicated available at https://www.cdc.gov/foodnet/reports/index.html. For the EU, data were obtained from The European Food Safety Authority for the year indicated at www.efsa.europa.eu/en/efsajournal/doc/ under reports entitled “The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in YYYY”, where YYYY indicates the year of interest.
In this study, a collection of mouse samples isolated during the mid- to late-1990s that was kept under monitored storage conditions was resuscitated to obtain a baseline of serotypes that were present at a time of peak egg contamination. The current study applied methodology for serotyping that is reliable for selecting and cataloguing strains for whole genome sequencing13). This study provides a historical reference for improving the quality of whole genome databases for the purpose of assessing evolutionary trends occurring in Salmonella enterica.
Live caught mice were obtained between March 1995 to May 1998 from 21 independent farm operators and four collective farm operators with 11 farms, for a total of 32 farms. All farms were primarily in Pennsylvania, although some bordering state lines may have been crossed. The 32 farm operators, who raised chicks to maturity, were supplied with birds from 7 different integrators, who supplied chicks for rearing. Egg production facilities housing more than 80,000 hens were included, and trapping of mice was part of implementing and assessing quality assurance programs. Farm data were coded, and no participant receiving samples had knowledge of the identity of the farm other than that farms were located in the Northeast region of the United States. Mice were euthanized by cervical dislocation and samples were collected aseptically, first spleens and then intestines. Samples were kept on ice and shipped to the US National Poultry Research Center (US NPRC), Athens, GA, USA for further analysis. Speciation of mice was by phenotypic characteristics, and rats were excluded. Thus, there is some possibility that mouse species other than Mus musculus may have been collected. Oversight of activities conducted on-farm, including collection of mice, was overseen by the Pennsylvania Department of Agriculture according to regulations in effect during the time period referenced.
2-2. Processing of Spleens and Intestines for SalmonellaSpleens were collected in 2 mL O-ring cryovials containing twenty 1 mm beads and 500 µL of Brain Heart Infusion Broth (BHI) (Difco, Becton-Dickinson, Franklin Lakes, NJ, US). Samples were placed in a bead beater 96-well high-throughput apparatus (Biospec, Bartlesville, OK, USA) for 3 minutes. A volume of 100 µL of suspension was plated onto Brilliant Green Plates (BG) (Neogen, Lansing, MI, US), and incubated for 24hr at 37°C. Colonies suspected of being Salmonella were then inoculated into Enterotube II (BBL, Becton-Dickinson), and 5 colonies per plate were processed. Positive cultures were frozen in 15% glycerol for storage at −80°C in monitored ultralow freezers. Mouse intestines were collected in 15 mL polypropylene centrifuge tubes with screw caps, 10 mL of BHI was added post-shipment, and then incubated at 48hr at 37°C. After the initial incubation, 1 mL was transferred to 10 mL Rappaport-Vassiliadis (RV) enrichment broth (Neogen) and incubated for 24hr at 37°C. Culture was streaked onto BG plates and incubated for 24hr at 37°C. If Salmonella colonies appeared to be mixed with other intestinal bacteria, multiple colonies were picked and then streaked again on BG as needed. As with spleens, at least 5 colonies were processed per intestinal sample, which were also frozen in 15% glycerol at −80°C. Revival of cultures was done by streaking frozen stock onto BG plates and incubating between 24 to 48hr at 37°C.
2-3. Determination of SerotypeSerotype and genotype (Serotype_genotype) were determined using dkgB-linked intergenic sequence ribotyping (ISR) and DNA microarray hybridization (DNAhyb) (Check & Trace Salmonella, Check-points BV, Wageningen)13,14). These methods correlate to the Kauffman-White antisera-based scheme15). Five colonies per isolate were processed to detect if multiple serotypes were present. Cells were pelleted from 1 mL BHI and DNA was extracted with the PureLInk Genomic DNA Mini Kit (Invitrogen, Life Technologies, Carlsbad, CA). One µl of DNA was added to 2X Gene Amp Fast PCR Master Mix (Applied Biosystems, Foster City, CA). The dkgB region was targeted by using amplification primers (Forward 5′ GCCAATGGCACTG CCCGGTA 3′; Reverse 5′ TACCGTGCGCTTTCGCCCAG 3′) within the same reaction, 200nM each. The final volume was adjusted to 30 µL. PCR was performed on a Veriti 96 well Fast Thermal cycler (Applied Biosystems Inc., Life Technologies) using the following conditions: 95°C for 10s, 35 cycles at 94°C, 40s at 64°C, and 10s at 72°C. The predicted amplicon of approximately 1400bp was confirmed by gel electrophoresis. PCR products were then purified using the QIA-quick PCR purification kit (Qiagen, Valencia, CA), and DNA concentrations were measured on a Synergy HT Multi-Detection microplate reader (Bio-Tek, Winooski, VT) at 260 nm. Sequencing primers (Forward 5′ AGGCCGGGTGTGTAAGCGCA 3′; Reverse CGGAACGGACGGGACTCGA 3′) were added to the PCR products for Sanger sequencing (Retrogen Inc., San Diego, CA) on a 3730 DNA Analyzer (Applied Biosystems Inc., Life Technologies). Mixtures of serotypes were observed as either different forward/reverse results for a single colony, or as multiple results for the 5 colonies processed. For determination of serotype by DNAhyb, any ISR result that appeared to be a mixture of serotypes was then further analyzed from large well-isolated colonies using Salmonella Check & Trace (Check-Points BV, Wageningen, The Netherlands)14).
2-4. Processing of ISR Raw Sequence DataAmbiguous base pairs (Ns) were trimmed from the 5′ and 3′ end of the raw sequence files. Forward and reverse sequences had different patterns of N distribution. For example, 60bp was usually trimmed from the 5′ ends in both forward and reverse directions, although some sequences required trimming 120bp. The 3′ ends of sequence files required different trimming strategies depending on direction. For the Forward direction, an average of 389 bp was trimmed to remove all Ns. For the Reverse direction, an average of 61 bp was trimmed. The range of sequence size that was used to search the ISR database and to conduct BLAST analysis of NCBI Salmonella enterica subspecies I was approximately between 540 − 660 bp. It is not recommended to use N-trimmed sequences of less than 300 bp for searches without review for quality of reaction. Sequence data were saved as Editseq files (DNAStar Lasergene 12 software package), and each file was approximately 1 − 2 kb. Parameters for conducting searches were 100 for Minimum Match Percentage and 100 for Minimum Sequence Length of 100 (Seqman Pro 12.0.0, DNASTAR, INC). The option for classic assembly was selected. Preassembly parameters were then set to allow no further addition of contigs prior to entry of test sequences from mice. Trimmed sequences for isolates in both directions were added and assembled. Only perfect matches were acceptable to assign serotype. A FASTA formatted table of the database is available upon request, and it currently includes 209 ISR sequences. Each unique ISR found within the set of mouse isolates was blasted against available microbe sequences at NCBI, and one accession number was recorded as representative of the ISR match if it was perfect. Serotypes Enteritidis and Typhimurium were searched individually by taxid.
Of 821 mice cultured, 157 were positive for Salmonella enterica (19.1%) (Table 1). Other information in Table 1 shows that the median sample group size was 30 mice per farm, average sample size submitted was 25.7 with a range from 3 − 55 per farm. The average number of positive mice per farm was 4.9, the median was 3.5, and the standard deviation was 6.9. Sample dates were, on average, 2 months apart, but farming schedules required flexibility. Therefore, the minimum sampling interval was 1 month, the maximum was 5 months, and the mode was 1 month. Of the 32 farms sampled, 21 (65.6%) were positive for Salmonella enterica subspecies I, and values ranged from 5.56% to 70% positive mice. Individual farm operator results ranged from 8.2 to 47.5% positive mice (Table 1). Of the 1,642 spleens and intestines cultured from all mice, 190 samples were positive (11.6%) (Table 1).
| Farm ID | Date received | Number of mice processed | Number of positive mice | Number of positive samples | Percent positive mice | Strain accession number3 |
| R040-01 | Jan-97 | 19 | 5 | 6 | 26.3% | 21025–21030 |
| R020-02 | Dec-95 | 10 | 5 | 5 | 50.0% | 20002–20006 |
| R010-02 | Jun-95 | 55 | 33 | 41 | 60.0% | 99053–99095 |
| R017-01 | Oct-95 | 34 | 13 | 13 | 38.2% | 99181–99193 |
| R009-02 | Jun-95 | 19 | 4 | 4 | 21.1% | 99049–99052 |
| R031-01 | Jun-96 | 30 | 0 | 0 | 0.0% | none isolated |
| R016-03 | Oct-95 | 13 | 8 | 13 | 61.5% | 99157–99162, 99174–99180 |
| R&Q052-06 | Dec-97 | 50 | 0 | 0 | 0.0% | none isolated |
| R043-02 | Jun-97 | 3 | 0 | 0 | 0.0% | none isolated |
| R032-01 | Jul-96 | 30 | 8 | 8 | 26.7% | 20117–20124 |
| R019-05 | Dec-95 | 30 | 2 | 2 | 6.7% | 20000–20001 |
| R036-02 | Oct-96 | 30 | 4 | 6 | 13.3% | 20159–20164 |
| R048-03 | Oct-97 | 30 | 2 | 2 | 6.7% | 21167–21168 |
| R015-01 | Sep-95 | 30 | 5 | 6 | 16.7% | 99148–99153 |
| R030-04 | May-96 | 30 | 21 | 27 | 70.0% | 20052–20078 |
| R051-02 | Dec-97 | 21 | 0 | 0 | 0.0% | none isolated |
| R005-03R | Mar-95 | 6 | 0 | 0 | 0.0% | none isolated |
| R027-01 | Apr-96 | 30 | 0 | 0 | 0.0% | none isolated |
| R012-01 | Jul-95 | 3 | 1 | 1 | 33.3% | 99111 |
| R042-02 | Feb-97 | 30 | 6 | 11 | 20.0% | 21041–21045, 21046–21051 |
| R029-06 | May-96 | 30 | 8 | 8 | 26.7% | 20042–20049 |
| R050-A&B | Dec-97 | 39 | 6 | 6 | 15.4% | 21169–21172, 22000, 22001 |
| R034-02 | Sep-96 | 30 | 9 | 11 | 30.0% | 20143–20153 |
| R028-01 | May-96 | 30 | 8 | 11 | 26.7% | 20031–20041 |
| R046-02 | Sep-97 | 3 | 0 | 0 | 0.0% | none isolated |
| R047-07 | Sep-97 | 9 | 0 | 0 | 0.0% | none isolated |
| R018-01 | Nov-95 | 30 | 0 | 0 | 0.0% | none isolated |
| R053-01 | Jan-98 | 24 | 0 | 0 | 0.0% | none isolated |
| R039-02 | Jan-97 | 30 | 0 | 0 | 0.0% | none isolated |
| R033-02 | Aug-96 | 30 | 4 | 4 | 13.3% | 20132–20135 |
| R041-01 | Feb-97 | 9 | 2 | 2 | 22.2% | 21039–21040 |
| R056-01&02 | May-98 | 54 | 3 | 3 | 5.6% | 22072–22074 |
| TOTALS | --- | 821 | 157 | 190 | --- | --- |
| Average | --- | 25.66 | 4.91 | 5.94 | --- | --- |
| Stdev | --- | 13.66 | 6.92 | 8.68 | --- | --- |
1Farm ID precedes the hyphen, and producer-specified information such as house number is after the hyphen.
2Only farms that had at least 3 mice submitted were included in analysis.
3Placing “USNPRC_” in front of the strain accession number, e. g. USNPRC_21025, will enable searching for genome information at NCBI (https://www.ncbi.nlm.nih.gov) 21).
Seven of the 33 mice with both positive spleens and intestines were carriers of more than one serotype, thus 164 isolations of serotypes were made from 157 different mice (Table 2). Other serotypes besides Enteritidis and Typhimurium that were isolated more than 10 times out of a total of 1,642 samples processed were Heidelberg and Schwarzengrund (Table 2). Serotypes Braenderup, Agona, and Manhattan were isolated fewer than 10 times each (Table 2). Serotypes Enteritidis (genovars 2994G and 2850), Heidelberg (genovar 15835), and Typhimurium (genovars 10909 and 2717) together accounted for 139 isolations, or 84.8% of all isolates.
| O-antigen group | (ISR reference) serotype name_genovar | Spleen samples | Intestinal samples | Both samples | Mixed, from spleen | Mixed, from intestines | Total isolates |
| D1 | (127) Enteritidis_2994G | 22 | 21 | 13 | 4 | 0 | 60 |
| B | (131) Heidelberg_15835 | 6 | 24 | 0 | 0 | 3 | 33 |
| B | (147) Typhimurium_10909 | 7 | 11 | 3 | 0 | 0 | 21 |
| D1 | (58) Enteritidis_2850 | 5 | 3 | 7 | 1 | 1 | 17 |
| B | (13) Schwarzengrund_14909BE | 13 | 2 | 0 | 2 | 0 | 17 |
| B | (6) Typhimurium 1,4,[5],12,i-_2717 | 1 | 4 | 3 | 0 | 0 | 8 |
| C1 | (55) Braenderup_9610 | 0 | 1 | 0 | 0 | 3 | 4 |
| B | (119) Agona_7205 | 0 | 2 | 0 | 0 | 0 | 2 |
| C1 | (21) Manhattan_11706 | 2 | 0 | 0 | 0 | 0 | 2 |
| TOTAL | 56 | 68 | 26 | 7 | 7 | 164 |
1A total of 157 mice were positive, and figures reflect that 2 different serotypes were isolated from 7 mice (columns 4 and 5).
Of the 157 mice positive for Salmonella, results based on sample source were (i) 56 (35.7%) positive for spleens only, (ii) 68 (43.3%) positive for intestines only, and (iii) 26 (16.6%) positive for both spleens and intestines (Table 2). Overall, 150 of the 157 positive mice (95.5%) were carriers of only one serotype after sampling 5 colonies per positive sample source. For the 1,642 samples cultured, 89 (46.8%) of positives were from spleens and 102 (53.7%) were from intestines. Of the 157 positive mice, only 33 (21.0%) had both positive spleens and intestines. Thus, culturing each mouse twice increased detection because both samples were positive from the same animal only about 20% of the time.
Percent recoveries of each serotype from the 56 mice with only positive spleens were in descending order: (i) Enteritidis_2994G, 39.3%, (ii) Schwarzengrund_14909B, 23.2%, (iii) Typhimurium_10909, 12.5%; (iv) Heidelberg_15835, 10.7%; (v) Enteritidis_2850, 8.9%; (vi) Manhattan_11706, 3.6%, and (vii) Typhimurium_1,4,[5],12a;I,-_2717, 1.8%. Percent isolations of each serotype from the 68 mice with only positive intestines were (i) Heidelberg_15835, 35.3%, (ii) Enteritidis_2994G, 30.9%, and (iii) Typhimurium_10909, 16.2%, (iv) Typhimurium_1,4,[5],12:I,-_2717, 5.9%, (v) Enteritidis_2850, 4.4%, (vi) Schwarzengrund_14909BE and Agona_7205, each at 2.9% of samples, and (vii) Braenderup_9610, 1.5%.
Respective to results from spleens and intestines, Typhimurium_10909 had 38.9% and 61.1% positives and Enteritidis_2994G had 46.5% and 48.8% positives. In addition, Enteritidis_2850 appeared to have similar isolation from both sample types as compared to Enteritidis_2994G. Too few isolates of mono-flagellated Typhimurium_2712 were obtained to make an assessment. In contrast, 13 of the 15 (86.7%) isolates of Schwarzengrund_14909BE were isolated from spleens only, and 24 of the 30 (80%) Heidelberg_15835 isolates were from intestines only. These results suggest that serotypes Enteritidis and Typhimurium are similarly efficient at both colonization and at organ invasion, and other serotypes such as Schwarzengrund and Heidelberg might differ.
Multiple serotypes of Salmonella enterica subspecies I appeared to be prevalent in the house mouse Mus musculus caught on poultry farms during the mid- to late-190s. Serotypes Enteritidis_2994DG and Typhimurium_10909 were especially prevalent. The time period is important because human salmonellosis that correlated to egg contamination by serotype Enteritidis was close to peak incidence, and biosecurity management was in the process of increasing. By 2004, illness in people had subsided to current levels, which is approximately 15 per 100,000 people in the United States (Fig. 1). It is possible that rodents had declined by 2004 following improvements in rodent control6). In the Garber study, 3.7% of 129 mice collected from 5 different regions of the US were positive for serotype Enteritidis, and the range was from 0 to 17.2%6). In this study, 19.1% of 821 mice had a range of serotypes, and almost half (49%) were positive for Enteritidis.
Results from this study highlight that mice can carry a variety of Salmonella enterica serotypes associated with foodborne illness. With the exception of serotype Manhattan, 8 of the 9 serotypes isolated are currently included within the top 30 serotypes commonly associated with foodborne salmonellosis16). The two serotypes most often associated with foodborne salmonellosis, namely Typhimurium and Enteritidis, were similarly isolated from both spleens and intestines. In contrast, other serotypes that are also important causes of foodborne illness might vary in mouse tissues. For example, Heidelberg and Schwarzengrund could have differences in colonization and invasive potential that should be considered when vaccinating poultry and when selecting a sampling strategy17).
Risk assessments have identified mice as impacting the safety of food6,8,11,18). Some guidance about the power of analysis required for future assessments is suggested. For example, collecting 113 mice from a farm in a single sampling appears adequate for assessing serotype prevalence if assumptions are that 5% of mice will have one serotype per animal at 99% confidence level, a margin of error of 5%, and an estimated population size of 1000 mice per location (http://www.raosoft.com/samplesize.html). If prevalence is dropped to 2% positive mice, results support that collecting even 50 naturally infected wild mice from a single farm could facilitate following trends associated with foodborne outbreaks of Salmonella enterica19). However, a rodent index of less than 20 was correlated with less Salmonella in poultry and ideally it should be difficult to recover 50 live mice as populations drop6). It is suggested that monitoring local mouse populations could facilitate protecting poultry from Salmonella. For example, farms experiencing a sudden increase in mouse populations could pursue serotyping of Salmonella of mice to evaluate vaccination strategies for reducing colonization of poultry. Whole-genome analysis will be required to determine detailed genome content of serotypes and to provide further information for assessing evolutionary trends and for selecting vaccine strains20). Sequencing and assembly of isolates are in progress with 64 isolates of serotype Enteritidis from this study currently available as unassembled reference sequences21).
This research was supported by the US Department of Agriculture in-house appropriation 6040–32000-011–00-D.
The authors have no conflict of interest.