ABSTRACT
In plant reproduction, pollination is the initial key process in bringing together the male and female gametophytes. When a pollen grain lands on the surface of the stigma, information is exchanged between the pollen and stigmatic cell to determine whether the pollen grain will be accepted or rejected. If it is accepted, the stigmatic papilla cell supplies water and other resources to the pollen for germination and pollen tube elongation. Cellular processes involving actin are essential for pollen germination and tube growth, and actin-binding proteins regulate these processes by interacting with actin filaments to assemble cytoskeletal structures and actin networks. LIM proteins, which belong to a subfamily of cysteine-rich proteins, are a family of actin-binding proteins in plants, and are considered to be important for formation of the actin cytoskeleton and maintenance of its dynamics. Although the physiological and biochemical characteristics of LIMs have been elucidated in vitro in a variety of cell types, their exact role in pollen germination and pollen tube growth during pollination remained unclear. In this manuscript, we focus on the pollen-specific LIM proteins, AtPLIM2a and AtPLIM2c, and define their biological function during pollination in Arabidopsis thaliana. The atplim2a/atplim2c double knockdown RNAi plants showed a reduced pollen germination, approximately one-fifth of wild type, and slower pollen tube growth in the pistil, that is 80.4 μm/hr compared to 140.8 μm/hr in wild type. These defects led to an occasional unfertilized ovule at the bottom of the silique in RNAi plants. Our data provide direct evidence of the biological function of LIM proteins during pollination as actin-binding proteins, modulating cytoskeletal structures and actin networks, and their consequent importance in seed production.
INTRODUCTION
Actin is an abundant protein in eukaryote cells. Actin monomers polymerize into actin filaments (AFs) to form the core element of the cytoskeleton and the tracks for intracellular signaling networks (Volkmann and Baluska, 1999; Drobak et al., 2004; Hamada et al., 2012). It has a crucial role in many cellular processes during the life of a cell, and it brings about changes in cell structure in response to developmental and environmental factors (Hussey et al., 2006; Staiger et al., 2010; Mugnai et al., 2012). To establish and maintain such cellular functions, a highly sophisticated system is required to regulate the organization and dynamics of the actin cytoskeleton. Regulation is carried out by actin-binding proteins (ABPs) and important subsets of ABPs are actin bundlers, which bind directly AFs to assemble cytoskeletal structures (actin bundles) and actin networks. ABP activities are regulated in association with many cellular parameters, such as Ca2+, pH and phosphorylation, to fulfill cellular requirements (Huang et al., 2005; Yokota et al., 2005; Papuga et al., 2010; Wen et al., 2012).
In higher plants, pollen grains germinate on the stigma and the emerging pollen tubes grow to the ovules for fertilization to complete sexual reproduction (Gutierrez-Marcos and Dickinson, 2012; Osaka et al., 2013). This process involves many actin-dependent cellular functions, such as dynamics of AFs, vesicle transport, endocytosis/exocytosis and formation of the cell wall (Cheung and Wu, 2008). In particular, the dynamics and organization of AFs are considered to be essential components of pollen germination and pollen tube growth, because suppression of AF organization by chemical treatment with latrunculin B or jasplakinolide results in disruption of pollen germination and pollen tube growth (Cárdenas et al., 2005; Chen et al., 2007; Lovy-Wheeler et al., 2007), suggesting that actin is essential for morphogenesis and cell elongation in plant reproduction (Kojo et al., 2013). ABPs regulate polymerization and depolymerization of AFs, and four ABP families, formin, villin, fimbrin, and LIM proteins, have been identified in plants (Thomas et al., 2009). These proteins are thought to regulate the organization, stabilization and dynamics of AFs collectively to assemble actin networks for cellular functions (Hussey et al., 2006; Staiger and Blanchoin, 2006). In Arabidopsis, Formin3 (Ye et al., 2009) and VILLIN5 (Zhang et al., 2010) are associated with the stabilization and organization of AFs in pollen. FIMBRIN1 in lily (Ll-FIM1) stabilizes the actin fringe (an actin structure in the subapex of the pollen tube) by cross-linking AFs in vitro, and injection of Ll-FIM1 antibody into pollen tubes resulted in slower tube growth compared to non-injected tubes (Su et al., 2012). Hence, it is clear that ABPs are important factors for assembly of the actin cytoskeleton during pollen germination and pollen tube growth.
LIM proteins (LIMs) function as ABPs in plants (Thomas et al., 2009), and are defined as relatively short proteins (approximately 200 amino acids) with two double zinc-finger LIM domains (40 to 50 amino acids), which function in protein-protein interaction. They belong to a subfamily of cysteine-rich proteins (CRPs) (Schmeichel and Beckerle, 1997; Weiskirchen and Gunther, 2003). LIMs are further classified into two subclasses, WLIMs and PLIMs. LIM genes belonging to the WLIM subclass are expressed throughout the sporophytic tissues in various organs, whereas LIM genes belonging to the PLIM subclass are expressed exclusively or preferentially in pollen (Baltz et al., 1999; Mundel et al., 2000). Tobacco WLIM1 has been shown to bind directly to AFs and triggers the regulation of actin cytoskeleton organization (Thomas et al., 2006). HaPLIM1 in sunflower co-localizes with AFs in the germination cone of the pollen grain during germination (Baltz et al., 1999). In lily, the in vitro interaction between pollen-enriched LIM1 (LILIM1) and AFs is regulated by pH and Ca2+, and it is suggested that this plays a central role in the oscillation of actin cytoskeleton remodeling during pollen tube elongation (Wang et al., 2008). In the Arabidopsis genome there are six LIM genes, three of which are classified as WLIMs (WLIM1, WLIM2a and WLIM2b) and the other three as PLIMs (PLIM2a, PLIM2b and PLIM2c). AtWLIMs are expressed widely throughout all sporophytic organs, including root, stem, leaves and flower. By contrast, AtPLIM2a and AtPLIM2c are expressed specifically in pollen and AtPLIM2b is expressed in the whole plant in addition to pollen (Arnaud et al., 2007; Ye and Xu, 2012). Expression levels of these PLIM2s in pollen increased at tri-cellular to maturation stages of pollen development (Papuga et al., 2010). PLIM2s co-localize with AFs in the pollen tube and direct actin-binding activity of PLIM2s has been demonstrated in vitro. Reduced pollen germination and shorter pollen tubes in vitro were observed in the triple mutants (AtPLIM2c: knockout, AtPLIM2a and AtPLIM2b: knockdown) (Ye and Xu, 2012). Because different distribution patterns of PLIM proteins were observed for PLIM2a and PLIM2c (that is longitudinally parallel actin bundles in the shank and short bundles in the sub-apical region for PLIM2a and long and short actin bundles in the germination pore of the pollen grain and in the tube shank but not in tube tip for PLIM2c) (Papuga et al., 2010; Ye et al., 2013), it is suggested that each PLIM protein has a distinct function in AF formation in pollen germination and pollen tube growth. Although biochemical analysis and in vitro pollen assays suggested a close relationship between PLIM2s and AFs in pollen germination and pollen tube growth (Ye and Xu, 2012), a direct confirmation of the biological function of PLIMs in pollen was not clear.
In this manuscript, we report the biological function of PLIM2s during pollination. Our results showed a reduction in pollen germination and slower pollen tube growth in the pistil in the double knockdown RNAi plants of AtPLIM2a and AtPLIM2c, leading to an occasional unfertilized seed at the bottom of the silique and a decreased total seed number per silique. Our data provide direct evidence of the function of PLIMs in pollen germination and tube growth during pollination and reveal the importance of their role in seed production in Arabidopsis.
MATERIALS AND METHODS
Plant materials
Arabidopsis thaliana ecotype Columbia (Col-0) plants were grown in a chamber (Panasonic-Sanyo, Tokyo, Japan) at 22℃ under 16 hr light/8 hr dark. Genomic DNA was extracted from fully expanded leaves of 6 to 8-week-old plants. Mature leaves, stems, roots, flower buds, and opened flowers were also collected for RT-PCR analysis.
RT-PCR analysis
Semiquantitative RT-PCR was performed according to Park et al. (2010) with slight modifications. Briefly, mRNA was isolated from the tissues described in Plant Materials using a Fast Track 2.0 mRNA isolation kit (Invitrogen, San Diego, CA, USA). First-strand cDNA was synthesized from 150 ng of isolated mRNA with a cDNA synthesis kit (First-Strand cDNA Synthesis kit, GE Healthcare, Piscataway, NJ, USA). The reverse-transcribed cDNA was used as a template for RT-PCR amplification with a specific primer set as listed in Supplementary Table S1. PCR was performed with ExTaq DNA polymerase (TaKaRa Bio, Shiga, Japan) for 35 cycles of denaturation for 1 min at 95℃, annealing for 1 min at 58℃ and extension for 1 min at 72℃, followed by a final extension for 4 min. As a positive control, a gene encoding actin4 (accession no. NM001085300) was used, as described in Hakozaki et al. (2008).
Construction of RNAi binary vectors
The 5’ upstream region containing putative promoter of AtPLIM2c was identified from the genomic sequence (At3g61230) of the Arabidopsis Database (TAIR: http://www.arabidopsis.org). A 1.10-kb DNA fragment from position –1 to –1,097 bp (the first ATG was marked as +1) was amplified by PCR using a primer set (AtPLIM2c-promoter-Forward and AtPLIM2c-promoter-Reverse), as described in Supplementary Table S1. A PCR fragment from the 5’ upstream region was subcloned into pCR2.1 plasmid vector (Invitrogen, San Diego, CA, USA). The nucleotide sequence was confirmed by sequencing of the plasmid using a ABI 310 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). The AtPLIM2c promoter was excised as a Kpn I-Sal I fragment from the plasmid and inserted into the Kpn I-Sal I site of the multicloning site in the binary vector pBI-GSH (Park et al., 2006), which contains a gene encoding β-glucuronidase (GUS; Ohta et al., 1990). This construct was named AtPLIM2c::GUS (Supplementary Fig. S1).
A binary vector AtPLIM2c::AtPLIM2c RNAi, having AtPLIM2c dsRNA interference (AtPLIM2c RNAi) governed by the AtPLIM2c promoter, was constructed. Sense and antisense 462-bp fragments of the 3’-region of AtPLIM2c from position 363 to 824 (with the first ATG marked as +1), containing the coding region and 3’-UTR, were amplified by PCR with a specific primer sets of AtPLIM2c-Sense-Forward and AtPLIM2c-Sense-Reverse, and AtPLIM2c-Antisense-Forward and AtPLIM2c-Antisense-Reverse, respectively (Supplementary Table S1). For the construction of a loop linker of RNAi vector (Chuang and Meyerowitz, 2000), the GUS coding region of 1,161 to 1,590 bp was amplified by the specific primer set of AtPLIM2c-GUS-Forward and AtPLIM2c-GUS-Reverse (Supplementary Table S1). To connect the above three PCR products, the following sequential PCRs were performed: PCR products of sense orientation of AtPLIM2c and GUS loop were connected by PCR with the primer set of AtPLIM2c-GUS-Forward and AtPLIM2c-sense-Reverse for GUS loop and AtLIM2c sense fragment (GUS-sense fragment), and PCR products of antisense orientation of AtPLIM2c and GUS loop were connected by PCR with the primer set of AtPLIM2c-antisense-Forward and AtPLIM2c-GUS-Reverse for AtLIM2c antisense and GUS loop fragment (antisense-GUS) (Supplementary Table S1). Then, the GUS-sense and antisense-GUS fragments were connected to make the antisense-GUS-sense fragment by PCR with primers of AtPLIM2c-Antisense-Forward and AtPLIM2c-Sense-Reverse (Supplementary Table S1). The antisense-GUS-sense fragment was subcloned into pCR2.1 vector and the nucleotide sequence was confirmed by sequencing, as described above. The antisense-GUS-sense plasmid was digested with Sal I and Sac I and subcloned downstream of the AtPLIM2c promoter in the AtPLIM2c::GUS binary vector, by replacing the GUS coding region. This RNAi construct was named AtPLIM2c::antisense-GUS-sense (Supplementary Fig. S1). The AtPLIM2c::antisense-GUS-sense was introduced into Agrobacterium tumefaciens strain EHA105 (Hood et al., 1993) by the electroporation method (Mersereau et al., 1990).
Plant transformation
Plant transformation was performed according to Park et al. (2006) and Park et al. (2010). Briefly, transgenic Arabidopsis plants were generated with the Agrobacterium binary vector by the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) containing 0.8% agar, 1% sucrose and 50 mg L–1 kanamycin. Transformants were grown in a growth chamber with conditions as described above.
Observation of pollen viability, pollen germination, and pollen tube growth
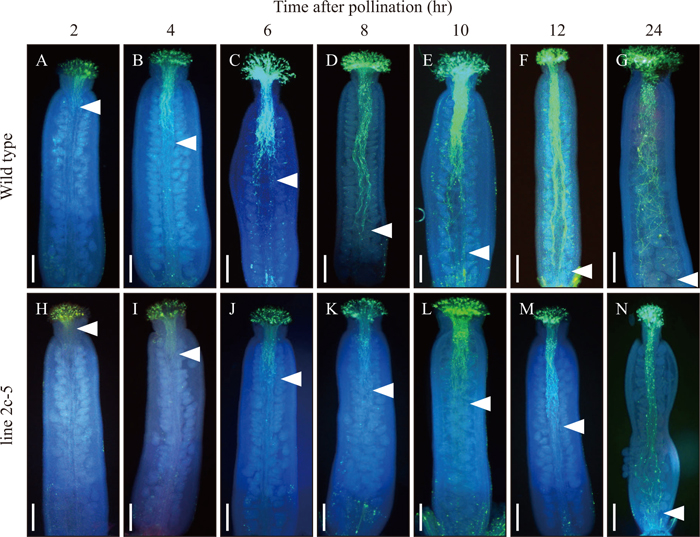
For observations of pollen viability at flowering stage 13 (Smyth et al., 1990), the Alexander staining method was performed (Alexander, 1969). Flowers at flowering stage 15 (Smyth et al., 1990) were collected from Arabidopsis plants and applied to an in vitro pollen germination assay. Pollen grains were placed on solid medium (pH7.5) containing 10% sucrose, 0.01% boric acid (w/v), 1 mM MgSO4, 5 mM CaCl2, 5 mM KCl, and 0.5% agar, at 22℃ for 6 hr (Boavida and McCormick, 2007). Germinated pollen grains and elongated pollen tubes were observed under a light microscope (Axio Imager A2, Carl Zeiss, Jena, Germany). For observations of pollen tube growth in vivo, pollinated pistils were collected every 2 hr, from 0 hr to 24 hr after pollination. Pistils were fixed in a 9:1 mixture of ethanol and acetic acid, stained with 0.1% aniline blue in 0.1% K3PO4 for 6 hr (Shimizu and Okada, 2000), and observed by UV fluorescence microscopy (Nikon, Eclipse E800 microscope system).
RESULTS
Construction of RNA interference plants of AtPLIM2a and AtPLIM2c
Of the three Arabidopsis PLIMs, AtPLIM2a and AtPLIM2c are expressed specifically in the pollen and AtPLIM2b is expressed in the whole plant in addition to pollen (Arnaud et al., 2007; Ye and Xu, 2012). Therefore in this study, we concentrated on elucidation of the role of pollen-specific AtPLIM2a and AtPLIM2c during pollination. RNA interference (RNAi) was used to investigate the biological functions of AtPLIM2s in pollination. In a previous report, an RNAi vector with the 5’ region of the AtPLIM2c (from 1 to 348 bp downstream of the ATG codon) targeted all three PLIM2 genes in transgenic plants, due to their high homology (Ye and Xu, 2012). Thus, to avoid this, RNAi plants specific for AtPLIM2a and AtPLIM2c were constructed using the 3’ region of the AtPLIM2c cDNA as an RNAi vector. As expected, transgenic RNAi plants showed decreased transcript levels of AtPLIM2a and AtPLIM2c compared to wild type Arabidopsis plants, while the transcript level of AtPLIM2b was unaffected. Two levels of repression were identified: severe (line 2c-5) and moderate (line 2c-39) (Fig. 1A). In both lines, there was no difference in plant growth and floral morphology compared to wild type (Fig. 1, B and C). However, some shorter siliques were found in line 2c-5 but not in line 2c-39 or wild type (Fig. 1C).

In order to investigate the shorter silique observed in line 2c-5, we evaluated pollen viability and silique content. Both wild-type and line 2c-5 showed normal pollen viability (Fig. 1B). However, the number of seeds per silique was different between wild type and line 2c-5 (Fig. 2 and Table 1): the average seeds per silique were 36.5 ± 7.3 in line 2c-5, compared to 47 ± 6.7 in wild type. This difference was significant in a t-test at 5% level. Furthermore, defective of seed development was frequently observed in the lower part of the silique in line 2c-5 (Fig. 2B). In reciprocal crosses between wild type and line 2c-5, defective seed development was observed (27.3 ± 5.3 seeds) when line 2c-5 was used as the pollen parent, but not as the pistil parent (41.5 ± 4.9 seeds) (Fig. 2A). The defect in seed development, in terms of number of seeds per silique and silique length, was commonly observed without respect to position of flowers in the inflorescence (Fig. 3). This result indicates that a repression of AtLIM2a and AtLIM2c affects pollen function in pollination.

Table 1. Silique length, number of seeds per silique and pollen germination in wild type and RNAi plants
| silique length
(mm) | No. of seeds* | pollen germination
(%)** |
|---|
| Wild type | 13.1 ± 1.80 | 47.0 ± 6.72 | 80.9 ± 3.56 |
| line 2c-5 | 12.1 ± 1.18 | 36.5 ± 7.26 | 16.2 ± 3.71 |
| line 2c-39 | 11.8 ± 0.87 | 41.8 ± 5.38 | 53.8 ± 5.81 |
Each value indicates mean ± SD. n = 30.
* Number of seeds per silique was significantly different between wild type and line 2c-5, and line 2c-5 and line 2c-39, respectively (p < 0.05).
** Pollen germination was significantly different between wild type and line 2c-5, wild type and line 2c-39, and line 2c-5 and line 2c-39, respectively (p < 0.05).

To characterize the defect in pollen function in line 2c-5, pollen germination and pollen tube growth were analyzed in vitro and in vivo, respectively. In line 2c-5 only 16.2% of pollen grains germinated, compared to 80.9% of pollen grains from wild type (Table 1). In addition, the reduction in percentage of pollen germinated was more severe in line 2c-5 than in line 2c-39. To observe pollen tube growth, pollinated pistils were fixed at 0, 2, 4, 6, 8, 10, 12, and 24 hr after pollination and observed by microscopy after aniline blue staining (Fig. 4). In the case of wild type, at 2 hr after pollination, pollen tubes were observed around the top part of the style, and at 12 hr after pollination pollen tubes had arrived at the basal part of the pistil, indicating that most pollen tubes reached the ovule for fertilization within 12 hr after pollination. In contrast, in line 2c-5, most pollen tubes were observed to be still in the stigma at 2 hr after pollination. At 6 hr after pollination, pollen tubes were in the upper quarter of the style, and at 12 hr after pollination pollen tubes were still around the middle part of the style, in contrast to the majority of pollen tubes in wild type, which had already arrived at the basal part of the style at this time. At > 24 hr after pollination, pollen tubes finally reached the basal part of style in line 2c-5. Based on these data, the rate of pollen tube growth was estimated at 140.8 μm/hr in wild type and 80.4 μm/hr in line 2c-5 (Fig. 5), and thus the rate of growth of pollen tubes in line 2c-5 was approximately 60% that of wild type. Taken together, these results indicate that reduced expression of AtPLIM2a and AtPLIM2c affects both pollen germination and pollen tube growth, resulting in shorter siliques with unfertilized ovules. The results also indicate that the level of reduction in expression of AtPLIM2a and AtPLIM2c directly correlated with the reduction in pollen germination and pollen tube growth.
DISCUSSION
Pollen tube growth is amazingly rapid compared to other growth processes in plants and involves extensive endo/exocytosis to bring about tip growth and rapid remodeling of the cellular cytoskeleton to support extension of the pollen tube. Among these biological functions, control of actin cytoskeleton dynamics is fundamental for the regulation of organelle movement, cytoplasmic streaming and vesicle transport (Hussey et al., 2006; Cárdenas et al., 2005; Suetsugu et al., 2012; Sugita et al., 2012; Kong et al., 2013). Four families of plant ABPs are thought to regulate formation of the actin cytoskeletal and maintenance of its flexibility (Thomas et al., 2009) and their physiological and biochemical characteristics have been elucidated in vitro in a variety of cell types (Yokota et al., 2005; Ye et al., 2009; Su et al., 2012). However, their exact contribution to pollen germination and pollen tube growth during pollination remained unclear, due to their functional complexity and gene redundancy (Ye and Xu, 2012).
LIM proteins are a family of ABPs in plants and their biochemical characteristics have been examined in tobacco, lily and Arabidopsis (Thomas et al., 2006; Wang et al., 2008; Papuga et al., 2010). WLIM1 in tobacco and LIM1 in lily directly bind to AFs and have been shown to modulate actin bundle assembly in cultured cells and in vitro pollen tubes. In Arabidopsis, the six LIM genes are categorized into two major groups by their expression pattern. Their expression patterns, however, are not specific but show considerable overlapped, and thus it is suggested that there is a high degree of functional redundancy. All Arabidopsis LIMs have been biochemically demonstrated to bind to AFs directly with different efficiencies (Papuga et al., 2010). LIM binding capabilities are differently regulated by pH and Ca2+: activities of all WLIMs are pH and Ca2+ independent while those of the three PLIM subclass members are pH dependent, being inhibited by high pH value (Papuga et al., 2010). In addition, activity of PLIM2c protein is down-regulated by high Ca2+, indicating that its activity is regulated by both pH and Ca2+. These characteristics suggest that Arabidopsis LIMs function in the same manner as other ABP proteins but also have specific functions in pollen.
In this study, pollen germination in plim2a/plim2c double knockdown RNAi plants was reduced significantly to 16.2 ± 3.7% in 2c-5 and 53.8 ± 5.8% in 2c-39, and correlated with the degree of reduction in expression of PLIM2a and PLIM2c (Fig. 1A and Table 1). Pollen tube growth was markedly delayed in plim2a/plim2c double knockdown plants and its speed was estimated to be 80.4 μm/hr, approximately 60% of that in wild type (Figs. 4 and 5). These reductions in pollen germination and pollen tube growth resulted in a failure of pollen tubes to access ovules in the lower part of pistils, leading to unfertilized ovules (Fig. 2B). Consequently, there was a reduction in the number of seeds set per silique and silique length (Figs. 2A, and 3, Table 1), which is as expected for a mutant defective in pollen tube growth (Jiang et al., 2005; Chae et al., 2009; Tanaka et al., 2013). In addition, these defects were observed only when the plim2a/plim2c double knockdown plants were used as the male parent, i.e. when pollens of the plim2a/plim2c double knockdown plants were pollinated onto stigmas of wild type or plim2a/plim2c double knockdown plants but not vice versa. Taken together, these results provide clear and direct evidence for PLIM function in pollen germination and pollen tube growth during pollination and our data support a central role of LIM proteins as ABPs in the regulation of AF organization during pollen germination and pollen tube growth.
In growing pollen tubes, gradients of pH and Ca2+ in the tip region are critical for correct elongation, and these factors are thought to be closely associated with actin cytoskeleton dynamics through the activation of ABPs. ABP activities themselves are also tightly regulated by many cellular parameters, including pH, Ca2+, phosphorylation, and protein-protein interactions (Yokota et al., 2005). For Arabidopsis pollen germination, a Ca2+ gradient in the pollen grain is required: thus, before pollen germination, Ca2+ increases at the potential germination site, presumably the germination pore, and pollen failing to establish a Ca2+ gradient does not germinate (Iwano et al., 2004). We observed defective pollen germination in plim2a/plim2c double knockdown RNAi plants (Table 1). Of the three PLIMs, only PLIM2c is responsive to Ca2+ concentration and is inactivated by high Ca2+ (Papuga et al., 2010), suggesting that modulation of actin organization by PLIM2c is involved in successful pollen germination. Our data support the proposition that each PLIM has a distinct role in pollen germination and pollen tube growth, although the three PLIMs are expressed together in Arabidopsis pollen grains. Papuga et al. (2010) reported that, in addition to expression in pollen grains, PLIM2a was observed in pollen tubes and sometimes in leaves, and PLIM2b was regularly observed in roots and leaf vascular tissues, but not in pollen tubes, while PLIM2c was restricted to pollen grains. In pollen tubes of the plim2a knockdown plants, AFs were irregular and distributed at random, and actin bundles in the subapical region of the tube shank were almost absent, leading to slower pollen tube elongation compared to that of wild type (Ye et al., 2013), while Ye and Xu (2012), suggested that reduced PLIM2a expression does not affect pollen germination. In the plim2a/plim2c double knockdown plants, phenotypic defects were observed in both pollen germination and pollen tube growth. From these results, taken all together, it is suggested that PLIM2a is the exclusive regulator of AF modulation during pollen tube growth, whereas actin dynamics for pollen germination is regulated by a cooperation of PLIM2a and PLIM2c (and possibly PLIM2b), indicating different roles for each PLIM during pollen germination and pollen tube growth.
This study shows the importance of PLIM2a and PLIM2c as ABPs and of their cooperative functions in pollination. However, the contribution of other ABPs in actin bundle assembly during pollination remains unclear. Therefore, their specific and individual functions in pollination, together with PLIMs, would be attractive candidates for further investigations to increase our understanding of pollination biology.
ACKNOWLEDGMENTS
The authors thank Masumi Miyano, Kana Ito, Momoko Ishikawa, Yoshie Higuchi, Nanase Kon, Ayako Tsushima, Reina Shida, and Ayumi Sadaike (Tohoku University) for technical assistance. This work was supported in part by Grants-in-Aid for Scientific Research on Innovative Areas (Nos. 23113006, 23113001 to G. S., K. S., and M. W.), Scientific Research (A) (No. 25252001 to M. W.), Scientific Research (B) (No. 25292005 to K. S.) and Scientific Research (C) (No. 25450515 to G. S.) from the Japan Society for Promotion of Science (JSPS). M. O. is recipient of a Research Fellowship for Young Scientists from JSPS.
REFERENCES
- Alexander, M. P. (1969) Differential staining of aborted and nonaborted pollen. Stain Technol. 44, 117–122.
- Arnaud, D., Déjardin, A., Leplé, J. C., Lesage-Descauses, M. C., and Pilate, G. (2007) Genome-wide analysis of LIM gene family in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa. DNA Res. 14, 103–116.
- Baltz, R., Schmit, A. C., Kohnen, M., Hentges, F., and Steinmetz, A. (1999) Differential localization of the LIM domain protein PLIM-1 in microspores and mature pollen grains from sunflower. Sex. Plant Reprod. 12, 60–65.
- Boavida, L. C., and McCormick, S. (2007) Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J. 52, 570–582.
- Cárdenas, L., Lovy-Wheeler, A., Wilsen, K. L., and Hepler, P. K. (2005) Actin polymerization promotes the reversal of streaming in the apex of pollen tubes. Cell Motil. Cytoskeleton 61, 112–127.
- Chae, K., Kieslich, C. A., Morikis, D., Kim, S. C., and Lord, E. M. (2009) A gain-of-function mutation of Arabidopsis lipid transfer protein 5 disturbs pollen tube tip growth and fertilization. Plant Cell 21, 3902–3914.
- Chen, T., Teng, N., Wu, X., Wang, Y., Tang, W., Samaj, J., Baluška, F., and Lin, J. (2007) Disruption of actin filaments by Latrunculin B affects cell wall construction in Picea meyeri pollen tube by disturbing vesicle trafficking. Plant Cell Physiol. 48, 19–30.
- Cheung, A. Y., and Wu, H. M. (2008) Structural and signaling networks for the polar cell growth machinery in pollen tubes. Annu. Rev. Plant Biol. 59, 547–572.
- Chuang, C. F., and Meyerowitz, E. M. (2000) Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97, 4985–4990.
- Clough, S. J., and Bent, A. F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743.
- Drobak, B. K., Frabklin-Tong, V. E., and Staiger, C. J. (2004) The role of the actin cytoskeleton in plant cell signaling. New Phytol. 163, 13–30.
- Gutierrez-Marcos, J. F., and Dickinson, H. G. (2012) Epigenetic reprogramming in plant reproductive lineages. Plant Cell Physiol. 53, 817–823.
- Hakozaki, H., Park, J. I., Endo, M., Takada, Y., Kazama, T., Takeda, Y., Suzuki, G., Kawagishi-Kobayashi, M., and Watanabe, M. (2008) Expression and developmental function of the 3-ketoacyl-ACP synthase2 gene in Arabidopsis thaliana. Genes Gent. Syst. 83, 143–152.
- Hamada, T., Tominaga, M., Fukaya, T., Nakamura, M., Nakano, A., Watanabe, Y., Hashimoto, T., and Baskin, T. I. (2012) RNA processing bodies, peroxisomes, golgi bodies, mitochondria, and endoplasmic reticulum tubule junctions frequently pause at cortical microtubules. Plant Cell Physiol. 53, 699–708.
- Hood, E. E., Gelvin, S. B., Melchers, L. S., and Hoekema, A. (1993) New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 2, 208–218.
- Huang, S., Robinson, R. C., Gao, L. Y., Matsumoto, T., Brunet, A., Blanchoin, L., and Staiger, C. J. (2005) Arabidopsis VILLIN1 generates actin filament cables that are resistant to depolymerization. Plant Cell 17, 486–501.
- Hussey, P. J., Ketelaar, T., and Deeks, M. J. (2006) Control of the actin cytoskeleton in plant cell growth. Annu. Rev. Plant Biol. 57, 109–125.
- Iwano, M., Shiba, H., Miwa, T., Che, F-S., Takayama, S., Nagai, T., Miyawaki, A., and Isogai, A. (2004) Ca2+ dynamics in a pollen grain and papilla cell during pollination of Arabidopsis. Plant Physiol. 136, 3562–3571.
- Jiang, L., Yang, S. L., Xie, L. F., Puah, C. S., Zhang, X. Q., Yang, W. C., Sundaresan, V., and Ye, D. (2005) VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell 17, 584–596.
- Kojo, K. H., Higaki, T., Kutsuna, N., Yoshida, Y., Yasuhara, H., and Hasezawa, S. (2013) Roles of cortical actin microfilament patterning in division plane orientation in plants. Plant Cell Physiol. 54, 1491–1503.
- Kong, S. G., Suetsugu, N., Kikuchi, S., Nakai, M., Nagatani, A., and Wada, M. (2013) Both phototropin 1 and 2 localize on the chloroplast outer membrane with distinct localization activity. Plant Cell Physiol. 54, 80–92.
- Lovy-Wheeler, A., Cárdenas, L., Kunkel, J. G., and Hepler, P. K. (2007) Differential organelle movement on the actin cytoskeleton in lily pollen tubes. Cell Motil. Cytoskeleton 64, 217–232.
- Mersereau, M., Pazour, G. J., and Das, A. (1990) Efficient transformation of Agrobacterium tumefaciens by electroporation. Gene 90, 149–151.
- Mugnai, S., Azzarello, E., Baluška, F., and Mancuso, S. (2012) Local root apex hypoxia induces NO-mediated hypoxic acclimation of the entire root. Plant Cell Physiol. 53, 912–920.
- Mundel, C., Baltz, R., Eliasson, A., Bronner, R., Grass, N., Kräuter, R., Evrard, J. L., and Steinmetz, A. (2000) A LIM-domain protein from sunflower is localized to the cytoplasm and/or nucleus in a wide variety of tissues and is associated with the phragmoplast in dividing cells. Plant Mol. Biol. 42, 291–302.
- Murashige, T., and Skoog, F. (1962) A revised medium for rapid growth and bio assay with tobacco tissue cultures. Physiol. Plant. 15, 473–497.
- Ohta, S., Mita, S., Hattori, T., and Nakamura, K. (1990) Construction and expression in tobacco of a β-glucuronidase (GUS) reporter gene containing an intron within the coding sequence. Plant Cell Physiol. 31, 805–813.
- Osaka, M., Matsuda, T., Sakazono, S., Masuko-Suzuki, H., Maeda, S., Sewaki, M., Sone, M., Takahashi, H., Nakazono, M., Iwano, M., et al. (2013) Cell type-specific transcriptome of Brassicaceae stigmatic papilla cells from a combination of laser microdissection and RNA sequencing. Plant Cell Physiol. 54, 1894–1906.
- Papuga, J., Hoffmann, C., Dieterle, M., Moes, D., Moreau, F., Tholl, S., Steinmetz, A., and Thomas, C. (2010) Arabidopsis LIM protein: a family of actin bundlers with distinct expression patterns and modes of regulation. Plant Cell 22, 3034–3052.
- Park, J.-I., Hakozaki, H., Endo, M., Takada, Y., Ito, H., Uchida, M., Okabe, T., and Watanabe, M. (2006) Molecular characterization of mature pollen-specific genes encoding novel small cysteine-rich proteins in rice (Oryza sativa L.). Plant Cell Rep. 25, 466–474.
- Park, J.-I., Ishimizu, T., Suwabe, K., Sudo, K., Masuko, H., Hakozaki, H., Nou, I.-S., Suzuki, G., and Watanabe, M. (2010) UDP-glucose phyrophosphorylase is rate limiting in vegetative and reproductive phases in Arabidopsis thaliana. Plant Cell Physiol. 51, 981–996.
- Schmeichel, K. L., and Beckerle, M. C. (1997) Molecular dissection of a LIM domain. Mol. Biol. Cell 8, 219–230.
- Shimizu, K. K., and Okada, K. (2000) Attractive and repulsive interactions between female and male gametophytes in Arabidopsis pollen tube guidance. Development 127, 4511–4518.
- Smyth, D. R., Bowman, J. L., and Meyerowitz, E. M. (1990) Early flower development in Arabidopsis. Plant Cell 2, 755–767.
- Staiger, C. J., and Blanchoin, L. (2006) Actin dynamics: old friends with new stories. Curr. Opin. Plant Biol. 9, 554–562.
- Staiger, C. J., Poulter, N. S., Henty, J. L., Franklin-Tong, V. E., and Blanchoin, L. (2010) Regulation of actin dynamics by actin-binding proteins in pollen. J. Exp. Bot. 61, 1969–1986.
- Su, H., Zhu, J., Cai, C., Pei, W., Wang, J., Dong, H., and Ren, H. (2012) FIMBRIN1 is involved in lily pollen tube growth by stabilizing the actin fringe. Plant Cell 24, 4539–4554.
- Suetsugu, N., Sato, Y., Tsuboi, H., Kasahara, M., Imaizumi, T., Kagawa, T., Hiwatashi, Y., Hasebe, M., and Wada, M. (2012) The KAC family of kinesin-like proteins is essential for the association of chloroplasts with the plasma membrane in land plants. Plant Cell Physiol. 53, 1854–1865.
- Sugita, C., Kato, Y., Yoshioka, Y., Tsurumi, N., Iida, Y., Machida, Y., and Sugita, M. (2012) CRUMPLED LEAF (CRL) homologs of Physcomitrella patens are involved in the complete separation of dividing plastids. Plant Cell Physiol. 53, 1124–1133.
- Tanaka, N., Uraguchi, S., Saito, A., Kajikawa, M., Kasai, K., Sato, Y., Nagamura, Y., and Fujiwara, T. (2013) Roles of pollen-specific boron efflux transporter, OsBOR4, in the rice fertilization process. Plant Cell Physiol. 54, 2011–2019.
- Thomas, C., Hoffmann, C., Dieterle, M., Van Troys, M., Ampe, C., and Steinmetz, A. (2006) Tobacco WLIM1 is a novel F-actin binding protein involved in actin cytoskeleton remodeling. Plant Cell 18, 2194–2206.
- Thomas, C., Tholl, S., Moes, D., Dieterle, M., Papuga, J., Moreau, F., and Steinmetz, A. (2009) Actin bundling in plants. Cell Motil. Cytoskeleton 66, 940–957.
- Volkmann, D., and Baluska, F. (1999) Actin cytoskeleton in plants: from transport networks to signaling networks. Microsc. Res. Tech. 47, 135–154.
- Wang, H. J., Wan, A. R., and Jauh, G. Y. (2008) An actin-binding protein, LlLIM1, mediates calcium and hydrogen regulation of actin dynamics in pollen tubes. Plant Physiol. 147, 1619–1636.
- Weiskirchen, R., and Gunther, K. (2003) The CRP/MLP/TLP family of LIM domain proteins: acting by connecting. Bioessays 25, 152–162.
- Wen, F., Wang, J., and Xing, D. (2012) A protein phosphatase 2A catalytic subunit modulates blue light-induced chloroplast avoidance movements through regulating actin cytoskeleton in Arabidopsis. Plant Cell Physiol. 53, 1366–1379.
- Ye, J., and Xu, M. (2012) Actin bundler PLIM2s are involved in the regulation of pollen development and tube growth in Arabidopsis. J. Plant Physiol. 169, 516–522.
- Ye, J., Zheng, Y., Yan, A., Chen, N., Wang, Z., Huang, S., and Yang, Z. (2009) Arabidopsis Formin3 directs the formation of actin cables and polarized growth in pollen tubes. Plant Cell 21, 3868–3884.
- Ye, J. R., Zhou, L. M., and Xu, M. L. (2013) Arabidopsis LIM proteins PLIM2a and PLIM2b regulate actin configuration during pollen tube growth. Biol. Plant. 57, 433–441.
- Yokota, E., Tominaga, M., Mabuchi, I., Tsuji, Y., Staiger, C. J., Oiwa, K., and Shimmen, T. (2005) Plant villin, lily P-135-ABP, possesses G-actin binding activity and accelerates the polymerization and depolymerization of actin in a Ca2+-sensitive manner. Plant Cell Physiol. 46, 1690–1703.
- Zhang, H., Qu, X., Bao, C., Khurana, P., Wang, Q., Xie, Y., Zheng, Y., Chen, N., Blanchoin, L., Staiger, C. J., and Huang, S. (2010) Arabidopsis VILLIN5, an actin filament bundling and severing protein, is necessary for normal pollen tube growth. Plant Cell 22, 2749–2767.





