2014 年 89 巻 4 号 p. 169-179
2014 年 89 巻 4 号 p. 169-179
Green fluorescent protein (GFP), fused to the N or C terminus of a protein of interest, is widely used to monitor the localization and mobility of proteins in cells. RAD51 is an essential protein that functions in mitotic DNA repair and meiotic chromosome segregation by promoting the homologous recombination reaction. A previous genetic study with Arabidopsis thaliana revealed that GFP fused to the C terminus of RAD51 (RAD51-GFP) inhibits mitotic DNA repair, but meiotic homologous recombination remained unaffected. To determine how the C-terminal GFP specifically inhibits mitotic DNA repair by RAD51, we purified rice RAD51A1-GFP and RAD51A2-GFP, and performed biochemical analyses. Interestingly, purified RAD51A1-GFP and RAD51A2-GFP are proficient in DNA binding and ATP hydrolysis. However, nucleoprotein complexes containing single-stranded DNA and RAD51A1-GFP or RAD51A2-GFP are significantly defective in binding to the second DNA molecule (secondary DNA binding), and consequently fail to catalyze homologous pairing. In contrast, RAD51A1-GFP and RAD51A2-GFP efficiently stimulated homologous pairing promoted by the meiosis-specific RAD51 isoform DMC1. These biochemical characteristics are well conserved in human RAD51-GFP. Therefore, GFP fused to the C terminus of RAD51 abolishes the homologous pairing activity of RAD51 by disrupting secondary DNA binding, but does not affect its DMC1-stimulating activity.
In meiotic cell division, homologous recombination is an essential process to ensure the proper segregation of homologous chromosomes (Petronczki et al., 2003; Neale and Keeney, 2006). Homologous recombination also functions in the repair of DNA double strand breaks (DSBs) in mitosis (Symington, 2002; West, 2003; San Filippo et al., 2008). During the homologous recombination process, homologous DNA regions between homologous chromosomes, sister chromatids or intrachromosomal repeat sequences are paired, in a reaction called homologous pairing (Symington, 2002; West, 2003; San Filippo et al., 2008; Renkawitz et al., 2014). In bacteria, RecA is the recombinase that catalyzes homologous pairing (Shibata et al., 1979; McEntee et al., 1979). In eukaryotes, two recombinases, RAD51 (denoted as Rad51 in Saccharomyces cerevisiae) and DMC1 (denoted as Dmc1 in S. cerevisiae), which are homologues of RecA (Aboussekhra et al., 1992; Basile et al., 1992; Bishop et al., 1992; Shinohara et al., 1992, 1993; Habu et al., 1996), catalyze homologous pairing (Sung, 1994; Baumann et al., 1996; Maeshima et al., 1996; Gupta et al., 1997; Li et al., 1997; Hong et al., 2001). The mammalian and yeast genomes contain one copy each of the RAD51 and DMC1 genes (Bishop et al., 1992; Shinohara et al., 1992, 1993; Habu et al., 1996). On the other hand, in the rice Oryza sativa japonica cultivar group, two non-allelic RAD51A1 and RAD51A2 genes, and two non-allelic DMC1A and DMC1B genes, have been identified (Ding et al., 2001; Shimazu et al., 2001; Kathiresan et al., 2002; Morozumi et al., 2013). Purified RAD51A1, RAD51A2, DMC1A and DMC1B promoted homologous pairing in vitro (Kant et al., 2005; Rajanikant et al., 2006, 2008; Sakane et al., 2008; Morozumi et al., 2013), suggesting that all four RAD51 and DMC1 proteins are bona fide recombinases in rice.
To promote the homologous pairing reaction in cells, RAD51 or DMC1 accumulates on single-stranded DNA (ssDNA) regions produced at DSB sites (primary DNA binding), and forms filamentous nucleoprotein complexes (Ogawa et al., 1993; Benson et al., 1994; Sung and Robberson, 1995; Sehorn et al., 2004; Renkawitz et al., 2014). The nucleoprotein complex then binds to double-stranded DNA (dsDNA) (secondary DNA binding), and forms a ternary complex containing ssDNA, dsDNA, and RAD51 or DMC1 (West, 2003; San Filippo et al., 2008; Renkawitz et al., 2014). Sequence homology between ssDNA and dsDNA is found in this ternary complex, and new base pairs are established between the invading ssDNA and the complementary strand of dsDNA, in an ATP-dependent manner.
Both RAD51 and DMC1 possess homologous pairing activity; however, they have distinct functions (Masson and West, 2001; Neale and Keeny, 2006; Kagawa and Kurumizaka, 2010; Murayama et al., 2011). RAD51 is produced in both mitotic and meiotic cells (Shinohara et al., 1992, 1993), whereas DMC1 is present only in meiotic cells (Bishop et al., 1992; Habu et al., 1996). Mitotic homologous recombinational DNA repair may depend on RAD51 function (Shinohara et al., 1992; Tashiro et al., 1996, 2000; Sonoda et al., 1998). On the other hand, the RAD51 and DMC1 recombinases are both required for meiotic homologous recombination (Bishop, 1994; Schwacha and Kleckner, 1997; Cloud et al., 2012; Da Ines et al., 2013; Lao et al., 2013; Liu et al., 2014). Recent genetic and biochemical studies with S. cerevisiae found that, in meiosis, Dmc1 plays the central role to promote homologous pairing, while Rad51 is down-regulated and required as an accessory factor for the Dmc1-mediated homologous pairing (Cloud et al., 2012; Liu et al., 2014).
Interestingly, in Arabidopsis thaliana, mitotic DNA repair reportedly becomes defective when green fluorescent protein (GFP) is fused to the C terminus of RAD51 (RAD51-GFP), but meiotic homologous recombination is not affected by the RAD51-GFP fusion (Da Ines et al., 2013). Similarly, human RAD51-GFP is reportedly defective in mitotic DNA repair, although when GFP is fused to the N terminus, RAD51 functions normally (Pellegrini et al., 2002; Yu et al., 2003; Forget et al., 2007). However, the mechanism by which the C-terminal GFP of RAD51 interferes with mitotic DNA repair, without affecting the meiotic function of RAD51, has not been clarified.
Here, to study the effect of the C-terminal GFP on the RAD51 activities, we purified rice RAD51A1-GFP, rice RAD51A2-GFP and human RAD51-GFP, and examined their recombinase activities in vitro. We selected rice RAD51A1 and RAD51A2 as representative plant RAD51 proteins, because they both share about 90% amino acid sequence identity with A. thaliana RAD51 and have been biochemically characterized (Morozumi et al., 2013). We then found that the GFP fusion to the C terminus of RAD51 did not affect the formation of the RAD51-ssDNA complexes, but specifically inhibited the secondary DNA binding of the RAD51-ssDNA complexes in both the rice and human RAD51 proteins. Interestingly, these rice and human RAD51-GFP proteins efficiently stimulated the DMC1-mediated homologous pairing in vitro. These results may explain why RAD51-GFP is specifically defective in mitotic DNA repair, but not in meiotic homologous recombination.
Rice (Oryza sativa) RAD51A1, rice RAD51A2, human RAD51, rice DMC1A and human DMC1 were prepared by methods described previously (Matsuo et al., 2006; Hikiba et al., 2008; Ishida et al., 2008; Sakane et al., 2008; Morozumi et al., 2013). For the RAD51-GFP proteins, the stop codons of rice RAD51A1, rice RAD51A2 and human RAD51 genes were replaced by a DNA fragment encoding Pro-Val-Ala-Thr as a linker peptide, and the enhanced GFP (EGFP) gene was fused just after the linker peptide sequence. EGFP is a single point mutant GFP derivative which exhibits enhanced fluorescence (Cormack et al., 1996). The resulting rice RAD51A1-GFP, rice RAD51A2-GFP and human RAD51-GFP genes were ligated into the NdeI-BamHI sites of the pET15b vector (Novagen, Darmstadt, Germany). Rice RAD51A1-GFP and RAD51A2-GFP were produced as N-terminally His6-tagged proteins, and were purified by the same method used for wild type RAD51A1 and RAD51A2 (Morozumi et al., 2013). In this method, the N-terminal His6-tag was proteolytically removed by thrombin protease (GE Healthcare Bio-Sciences, Uppsala, Sweden), and thus the purified rice RAD51A1-GFP and RAD51A2-GFP did not contain the His6-tag.
Human RAD51-GFP was produced as an N-terminally His6-tagged protein in the Escherichia coli BLR (DE3) pLysS strain, which also carried an expression vector for the minor tRNAs (Codon(+) RIL; Agilent Technologies, Santa Clara, USA). Cells producing the protein were resuspended in buffer A (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 5 mM imidazole, 5 mM 2-mercaptoethanol, 10% glycerol) and disrupted by sonication. Cell debris was removed by centrifugation. The supernatant was gently mixed with nickel-nitrilotriacetic acid (Ni-NTA) agarose beads (QIAGEN, Venlo, Netherlands) at 4℃ for 1 h, and the beads were washed with 50 column volumes (CVs) of buffer B (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10 mM imidazole, 5 mM 2-mercaptoethanol, 10% glycerol). Proteins bound to the Ni-NTA beads were eluted by a linear gradient of 10 to 500 mM imidazole. Fractions containing His6-RAD51-GFP were collected, and the His6-tag was removed by thrombin protease treatment (7 units/mg) during dialysis against buffer C (50 mM Tris-HCl, pH 8.0, 200 mM KCl, 0.25 mM EDTA, 2 mM 2-mercaptoethanol, 10% glycerol), after which the sample was loaded onto a Heparin Sepharose column (GE Healthcare). The column was washed with 50 CVs of buffer C, and the proteins were eluted by a linear gradient of 200 to 1,000 mM KCl. Peak fractions containing RAD51-GFP were dialyzed against buffer C, and the sample was further purified by MonoQ column chromatography (GE Healthcare). After the sample was loaded, the column was washed with 10 CVs of buffer C, and RAD51-GFP was eluted by buffer C containing 600 mM KCl. The purified RAD51-GFP protein was dialyzed against buffer D (20 mM HEPES-NaOH, pH 7.5, 150 mM NaCl, 0.1 mM EDTA, 2 mM 2-mercaptoethanol, 10% glycerol), and stored at –80℃.
DNA substratesSingle-strand ϕX174 viral (+) strand DNA and double-strand ϕX174 replicative form I DNA were purchased from New England Biolabs (Ipswich, USA). Linear dsDNA was prepared from the ϕX174 replicative form I DNA by PstI digestion. For ternary complex formation and D-loop formation assays, superhelical dsDNA containing tandem repeats of the 5S rDNA sequence was used. The superhelical dsDNA was prepared with a method avoiding irreversible denaturation by alkaline treatment of the cells (Kagawa et al., 2001). For the ssDNA substrates, the following oligonucleotides, purified by high-performance liquid chromatography, were purchased from Nihon Gene Research Laboratory (Sendai, Japan):
5S 90-mer, 5’ CCGGT ATATT CAGCA TGGTA TGGTC GTAGG CTCTT GCTTG ATGAA AGTTA AGCTA TTTAA AGGGT CAGGG ATGTT ATGAC GTCAT CGGCT 3’ and
5S 70-mer, 5’ CCGGT ATATT CAGCA TGGTA TGGTC GTAGG CTCTT GCTTG ATGAA AGTTA AGCTA TTTAA AGGGT CAGGG 3’.
DNA binding assayFor rice RAD51A1, RAD51A2, RAD51A1-GFP and RAD51A2-GFP, the reactions were performed in a reaction buffer containing 25 mM HEPES-NaOH, pH 7.5, 1 mM DTT, 3 mM ATP and 1 mM MgCl2. For human RAD51 and RAD51-GFP, the reactions were performed in a buffer containing 20 mM HEPES-NaOH, pH 7.5, 1 mM DTT, 1 mM ATP, 1 mM MgCl2, 2 mM CaCl2, 20 mM creatine phosphate and 75 μg/ml creatine kinase. The indicated amounts of rice RAD51A1, rice RAD51A2, rice RAD51A1-GFP, rice RAD51A2-GFP, human RAD51 and human RAD51-GFP were incubated with ϕX174 circular ssDNA (20 μM in nucleotides) or linearized ϕX174 dsDNA (20 μM in nucleotides) in the appropriate reaction buffer. The samples were incubated at 37℃ for 15 min, and were separated by 0.8% agarose gel electrophoresis in 1× TAE buffer at 3.3 V/cm for 2.5 h. Bands were visualized by ethidium bromide staining.
ATPase assayRice RAD51A1, rice RAD51A2, rice RAD51A1-GFP, rice RAD51A2-GFP, human RAD51 and human RAD51-GFP (1.5 μM each) were incubated at 37℃ for 30 min in the presence of ϕX174 circular ssDNA (20 μM in nucleotides), in 10 μl of reaction buffer containing 24 mM HEPES-NaOH, pH 7.5, 1 mM MgCl2, 0.02 mM EDTA, 80 mM NaCl, 0.4 mM 2-mercaptoethanol, 1 mM DTT, 2% glycerol, 5 μM ATP and 5 nCi [γ-32P]ATP (NEG502A, PerkinElmer, Waltham, USA). The reaction was quenched by the addition of 5 μl of 0.5 M EDTA, pH 8.0, and ATP, ADP and AMP were separated by thin layer chromatography on polyethyleneimine-cellulose, in a 0.5 M LiCl and 1 M formic acid solution.
Assays for ternary complex formation and D-loop formationFor rice RAD51A1, RAD51A2, DMC1A, RAD51A1-GFP and RAD51A2-GFP, the reactions were performed in a buffer containing 25 mM HEPES-NaOH, pH 7.5, 1 mM DTT, 3 mM ATP and 1 mM MgCl2. Human RAD51 and DMC1 reportedly require Ca2+ for efficient homologous pairing (Bugreev and Mazin, 2004; Bugreev et al., 2005). Therefore, for human RAD51, RAD51-GFP and DMC1, the reactions were performed in a buffer containing 20 mM HEPES-NaOH, pH 7.5, 1 mM DTT, 1 mM ATP, 1 mM MgCl2, 2 mM CaCl2, 20 mM creatine phosphate and 75 μg/ml creatine kinase. The indicated amounts of rice RAD51A1, rice RAD51A2, rice RAD51A1-GFP, rice RAD51A2-GFP, human RAD51, human RAD51-GFP, rice DMC1A and human DMC1 were incubated with a 32P-labeled single-stranded oligonucleotide (ssDNA) on ice for 15 min, and then at 37℃ for 5 min. Subsequently, 1 μl of superhelical plasmid DNA was added, and the reactions were continued at 37℃ for 10 min. For rice RAD51A1, RAD51A2, RAD51A1-GFP, RAD51A2-GFP and DMC1A, the reactions were conducted with 5S 70-mer ssDNA (3 μM in nucleotides) and superhelical dsDNA (30 μM in nucleotides) as the DNA substrates. For human RAD51, RAD51-GFP and DMC1, the reactions were conducted with 5S 90-mer ssDNA (3 μM in nucleotides) and superhelical dsDNA (100 μM in nucleotides) as substrates.
For the ternary complex formation assay, glutaraldehyde was added to a final concentration of 0.09%, and the samples were incubated for 10 min. Nucleoprotein complexes were separated by 1% agarose gel electrophoresis in 0.5× TBE buffer at 3.3 V/cm for 2.5 h. For the D-loop formation assay, the reactions were terminated by the addition of 2 μl of stop solution containing 0.2% SDS and 1.4 mg/ml proteinase K (Roche Applied Science, Indianapolis, USA), and were incubated at 37℃ for 15 min. The deproteinized DNA products were separated by 1% agarose gel electrophoresis in 1× TAE buffer at 4 V/cm for 2 h. The gels were dried, and gel images were obtained using an FLA-7000 imaging analyzer (Fujifilm, Tokyo, Japan). Band intensities were quantitated with Multi Gauge software (Fujifilm).
Rice RAD51A1-GFP and RAD51A2-GFP were bacterially produced as recombinant proteins, and were purified to near homogeneity (Fig. 1A). We first tested their DNA-binding activities by performing gel shift assays with circular ssDNA and linearized dsDNA. As shown in Fig. 1B and Fig. 1C, RAD51A1-GFP and RAD51A2-GFP bound to ssDNA and dsDNA as efficiently as the wild type RAD51A1 and RAD51A2 proteins. This ssDNA or dsDNA binding may occur at the same DNA-binding sites within the RAD51 filament (primary DNA binding).

DNA-binding and ATP-hydrolyzing activities of rice RAD51A1-GFP and RAD51A2-GFP. (A) Purified rice RAD51A1, RAD51A2, RAD51A1-GFP and RAD51A2-GFP (0.75 μg each) were analyzed by 12% SDS-PAGE with Coomassie Brilliant Blue staining. (B) ssDNA-binding activities of rice RAD51A1-GFP and RAD51A2-GFP. ϕX174 circular ssDNA (20 μM) was incubated with RAD51A1 (lanes 2–4), RAD51A2 (lanes 5–7), RAD51A1-GFP (lanes 8–10) or RAD51A2-GFP (lanes 11–13) at 37℃ for 15 min. The samples were separated by 0.8% agarose gel electrophoresis. The protein concentrations were 1 μM (lanes 2, 5, 8 and 11), 2 μM (lanes 3, 6, 9 and 12) and 4 μM (lanes 4, 7, 10 and 13). Lane 1 is a negative control experiment without proteins. css: circular single-stranded DNA. (C) dsDNA-binding activities of rice RAD51A1-GFP and RAD51A2-GFP. Linear ϕX174 dsDNA (20 μM) was incubated with RAD51A1 (lanes 2–4), RAD51A2 (lanes 5–7), RAD51A1-GFP (lanes 8–10) or RAD51A2-GFP (lanes 11–13) at 37℃ for 15 min. The samples were separated by 0.8% agarose gel electrophoresis. The protein concentrations were 1 μM (lanes 2, 5, 8 and 11), 2 μM (lanes 3, 6, 9 and 12) and 4 μM (lanes 4, 7, 10 and 13). Lane 1 is a negative control experiment without proteins. lds: linear double-stranded DNA. (D and E) ATP-hydrolyzing activities of rice RAD51A1-GFP and RAD51A2-GFP. The reactions were conducted in the presence (D) or absence (E) of ϕX174 circular ssDNA (20 μM) at 37℃ for 30 min. The average and standard deviation values of three independent experiments are shown.
RAD51 hydrolyzes ATP in the presence of ssDNA (Sung, 1994). As reported previously (Morozumi et al., 2013), RAD51A1 and RAD51A2 efficiently hydrolyzed ATP in the presence of ssDNA, but not in its absence (Fig. 1, D and E). Interestingly, RAD51A1-GFP and RAD51A2-GFP exhibited ATPase activities with ssDNA that were similar to those of the wild type proteins (Fig. 1D). In the absence of ssDNA, the ATP-hydrolyzing activities of RAD51A1-GFP and RAD51A2-GFP were extremely low (Fig. 1E). These results suggested that RAD51A1-GFP and RAD51A2-GFP are properly folded into tertiary structures, and are proficient in primary DNA binding to form the nucleoprotein complex, in which RAD51 hydrolyzes ATP.
Rice RAD51A1-GFP and RAD51A2-GFP are specifically defective in secondary DNA binding for ternary complex formationWe then tested the secondary DNA-binding activities of RAD51A1-GFP and RAD51A2-GFP, by the ternary complex formation assay. In the secondary DNA-binding step, the dsDNA must bind to sites that are different from the primary DNA-binding sites of the RAD51 filament, because the primary DNA-binding sites are occupied by the ssDNA. In the ternary complex formation assay, a 32P-labeled ssDNA 70-mer was first incubated with wild type RAD51A1, wild type RAD51A2, RAD51A1-GFP or RAD51A2-GFP, to form nucleoprotein complexes containing ssDNA (Fig. 2A). Superhelical dsDNA was then added to the reaction mixtures, and the resulting ternary complexes containing ssDNA, dsDNA and RAD51 were separated on an agarose gel, after crosslinking with glutaraldehyde (Fig. 2A). As shown in Fig. 2B and Fig. 2C (lanes 3–5), substantial amounts of ternary complexes were detected in the presence of wild type RAD51A1 and RAD51A2, as a consequence of secondary DNA binding by the RAD51A1-ssDNA and RAD51A2-ssDNA complexes. Interestingly, we found that both RAD51A1-GFP and RAD51A2-GFP were significantly defective in ternary complex formation. However, the amounts of the complexes with ssDNA were comparable to those of the complexes of wild type RAD51A1 and RAD51A2 (Fig. 2, B and C).
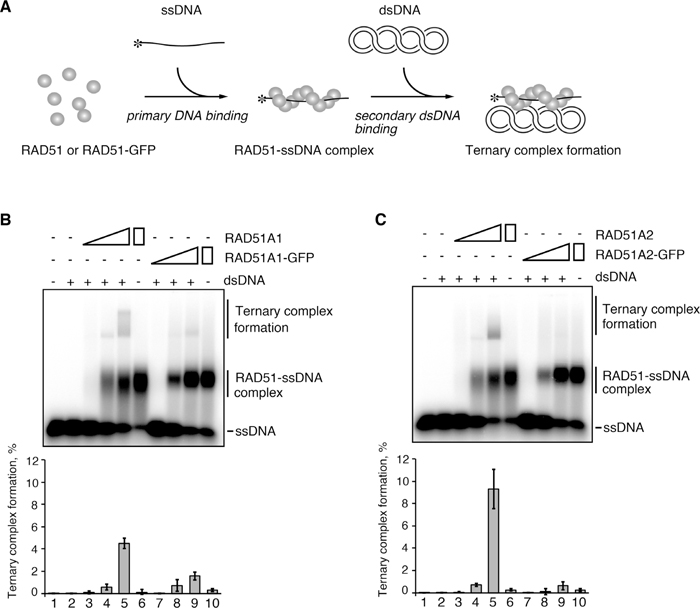
Rice RAD51A1-GFP and RAD51A2-GFP are defective in secondary DNA binding. (A) Scheme of the ternary complex formation assay. Asterisks indicate the 32P-labeled 5’-end of the ssDNA. (B and C) Rice RAD51A1 (B), RAD51A1-GFP (B), RAD51A2 (C) or RAD51A2-GFP (C) was incubated with 32P-labeled ssDNA (3 μM). The reactions were initiated by the addition of superhelical dsDNA (30 μM), and were continued at 37℃ for 10 min. The protein concentrations were 0.2 μM (lanes 3 and 7), 0.6 μM (lanes 4 and 8) and 1 μM (lanes 5, 6, 9 and 10). Lanes 1 and 2 are negative control experiments without proteins in the absence and presence of dsDNA, respectively. Lanes 6 and 10 are control experiments with RAD51A1 (B), RAD51A1-GFP (B), RAD51A2 (C) or RAD51A2-GFP (C) in the absence of dsDNA. The samples were separated by 1% agarose gel electrophoresis, after crosslinking with glutaraldehyde. The average and standard deviation values of three independent experiments are shown in the bottom panel.
Consistent with their defective secondary DNA-binding activities, RAD51A1-GFP and RAD51A2-GFP were markedly defective in homologous pairing activities, as evaluated by the D-loop formation assay (Fig. 3). In this assay, a 32P-labeled ssDNA 70-mer and superhelical dsDNA were used as DNA substrates, and D-loops, in which the 32P-labeled ssDNA pairs with the complementary strand of the dsDNA, were detected as the product by homologous pairing (Fig. 3A). As shown in Fig. 3B and Fig. 3C, the homologous pairing activities of RAD51A1-GFP and RAD51A2-GFP were drastically decreased relative to those of wild type RAD51A1 and RAD51A2. Therefore, we concluded that the GFP fused to the C terminus of RAD51A1 or RAD51A2 specifically interferes with the secondary DNA binding of the RAD51-ssDNA complex, and consequently inhibits homologous pairing.

Rice RAD51A1-GFP and RAD51A2-GFP are defective in homologous pairing. (A) Scheme of the D-loop formation assay. Asterisks indicate the 32P-labeled 5’-end of the ssDNA. (B and C) Rice RAD51A1 (B), RAD51A1-GFP (B), RAD51A2 (C) or RAD51A2-GFP (C) was incubated with 32P-labeled ssDNA (3 μM), and the reactions were initiated by the addition of superhelical dsDNA (30 μM) and continued at 37℃ for 10 min. The protein concentrations were 0.2 μM (lanes 2 and 5), 0.6 μM (lanes 3 and 6), and 1 μM (lanes 4 and 7). Lane 1 is a negative control experiment without proteins. The reactions were stopped by the addition of SDS and proteinase K, and the deproteinized DNA samples were separated by 1% agarose gel electrophoresis. The average and standard deviation values of three independent experiments are shown in the bottom panel.
In the plant A. thaliana, RAD51-GFP is defective in mitotic DNA repair, but is proficient in meiotic homologous recombination (Da Ines et al., 2013). This prompted us to hypothesize that RAD51-GFP may be defective in its homologous pairing activity required for mitotic DNA repair, but proficient in stimulation activity for meiotic DMC1-mediated homologous pairing.
Stimulation of DMC1-mediated homologous pairing by RAD51 has been observed in S. cerevisiae (Cloud et al., 2012). Therefore, we first tested whether rice RAD51A1 and RAD51A2 could stimulate homologous pairing mediated by rice DMC1A (Fig. 4A). To do so, we performed D-loop formation assays with a fixed amount of DMC1A, and increasing amounts of RAD51A1 or RAD51A2 were added to the reaction mixtures. We found that both RAD51A1 and RAD51A2 efficiently stimulated the homologous pairing promoted by DMC1A (Fig. 4B). These results suggested that the DMC1-stimulating activity of RAD51 is conserved between the yeast and plant systems.

Rice RAD51A1-GFP and RAD51A2-GFP stimulate rice DMC1A-mediated homologous pairing. (A) Purified rice DMC1A (0.75 μg) was analyzed by 12% SDS-PAGE with Coomassie Brilliant Blue staining. (B) Stimulation of DMC1A-mediated homologous pairing by rice RAD51A1 and RAD51A2. Rice DMC1A (1 μM) was incubated with 32P-labeled ssDNA (3 μM) in the presence of RAD51A1 or RAD51A2. The rice RAD51A1 and RAD51A2 concentrations were 0.2 μM (lanes 3 and 7), 0.4 μM (lanes 4 and 8) and 0.6 μM (lanes 5, 6, 9 and 10). The reactions were initiated by the addition of superhelical dsDNA (30 μM) and continued at 37℃ for 10 min. Lane 1 is a negative control experiment without proteins. Lane 2 is a control experiment without RAD51A1 or RAD51A2, in the presence of DMC1A. Lanes 6 and 10 represent control experiments with RAD51A1 (0.6 μM) and RAD51A2 (0.6 μM), in the absence of DMC1A. The reactions were stopped by the addition of SDS and proteinase K, and the deproteinized DNA samples were separated by 1% agarose gel electrophoresis. The average and standard deviation values of three independent experiments are shown in the bottom panel. (C) Stimulation of DMC1A-mediated homologous pairing by rice RAD51A1-GFP and RAD51A2-GFP. Rice DMC1A (1 μM) was incubated with 32P-labeled ssDNA (3 μM) in the presence of RAD51A1-GFP or RAD51A2-GFP. The rice RAD51A1-GFP and RAD51A2-GFP concentrations were 0.2 μM (lanes 3 and 7), 0.4 μM (lanes 4 and 8) and 0.6 μM (lanes 5, 6, 9 and 10). The experiments were performed as described in (B).
We next tested the DMC1-stimulating activity of RAD51A1-GFP and RAD51A2-GFP in homologous pairing. Intriguingly, both RAD51A1-GFP and RAD51A2-GFP significantly stimulated the homologous pairing mediated by DMC1A (Fig. 4C), although they were defective in their own homologous pairing activities (Fig. 3). These results indicated that rice RAD51A1-GFP and RAD51A2-GFP clearly have stimulatory activity for DMC1-mediated homologous pairing, which may play an essential role in meiotic homologous recombination.
Human RAD51-GFP has similar characteristics to rice RAD51A1-GFP and RAD51A2-GFPTo test whether the DMC1-stimulating activity of RAD51 is also conserved in mammals, we purified the human RAD51-GFP protein (Fig. 5A). Rice RAD51A1 and RAD51A2 share about 70% amino acid identity with human RAD51 (Morozumi et al., 2013). The purified human RAD51-GFP bound to ssDNA and dsDNA (Fig. 5, B and C), and hydrolyzed ATP in the presence of ssDNA (Fig. 5D). Like rice RAD51A1-GFP and RAD51A2-GFP, human RAD51-GFP was significantly defective in ternary complex formation and homologous pairing (Fig. 6, A and B), indicating that the GFP interferes with the secondary DNA binding of the RAD51-ssDNA complex. We found that human RAD51-GFP exhibited robust stimulation of DMC1-mediated homologous pairing, to a level comparable to wild type RAD51 (Fig. 6C). Therefore, GFP fused to the C terminus of RAD51 commonly inhibits secondary DNA binding during homologous pairing, without affecting RAD51-ssDNA complex formation, and retains DMC1-stimulating activity. We also concluded that the stimulatory function of RAD51 in DMC1-mediated homologous pairing is widely conserved among the yeast, plant and mammalian systems.

DNA-binding and ATP-hydrolyzing activities of human RAD51-GFP. (A) Purified human DMC1, RAD51 and RAD51-GFP (0.75 μg each) were analyzed by 12% SDS-PAGE with Coomassie Brilliant Blue staining. Lane 1 contains molecular mass markers. (B) ssDNA-binding activities of human RAD51-GFP. ϕX174 circular ssDNA (20 μM) was incubated with RAD51 (lanes 2–5) or RAD51-GFP (lanes 7–10) at 37℃ for 15 min. The samples were separated by 0.8% agarose gel electrophoresis. The protein concentrations were 0.5 μM (lanes 2 and 7), 1 μM (lanes 3 and 8), 2 μM (lanes 4 and 9) and 4 μM (lanes 5 and 10). Lanes 1 and 6 are negative control experiments without proteins. css: circular single-stranded DNA. (C) dsDNA-binding activities of human RAD51-GFP. The experiments were performed as described in (B), except that linear ϕX174 dsDNA (20 μM) was used instead of ssDNA. lds: linear double-stranded DNA. (D) ATP-hydrolyzing activity of human RAD51-GFP. The reactions were conducted in the presence of ϕX174 circular ssDNA (20 μM), at 37℃ for 30 min. The negative control experiment without proteins represents the same data presented in Fig. 1D. The average and standard deviation values of three independent experiments are shown.

Stimulation of human DMC1-mediated homologous pairing by human RAD51. (A) Ternary complex formation assay. Human RAD51 or RAD51-GFP was incubated with 32P-labeled ssDNA (3 μM), and the reactions were initiated by the addition of superhelical dsDNA (100 μM) and continued at 37℃ for 10 min. The protein concentrations were 0.125 μM (lanes 3 and 8), 0.25 μM (lanes 4 and 9), 0.5 μM (lanes 5 and 10) and 1 μM (lanes 6, 7, 11 and 12). Lanes 1 and 2 are negative control experiments without proteins in the absence and presence of dsDNA, respectively. Lanes 7 and 12 are control experiments with RAD51 and RAD51-GFP, respectively, in the absence of dsDNA. The samples were separated by 1% agarose gel electrophoresis, after crosslinking with glutaraldehyde. The average and standard deviation values of three independent experiments are shown in the bottom panel. (B) Homologous pairing activity of RAD51-GFP. Human RAD51 (lanes 2–5) or RAD51-GFP (lanes 6–9) was incubated with a 32P-labeled 90-mer oligonucleotide (3 μM), and the reactions were initiated by the addition of superhelical dsDNA (100 μM) and continued at 37℃ for 10 min. The protein concentrations were 0.125 μM (lanes 2 and 6), 0.25 μM (lanes 3 and 7), 0.5 μM (lanes 4 and 8) and 1 μM (lanes 5 and 9). Lane 1 is a negative control experiment without proteins. The reactions were stopped by the addition of SDS and proteinase K, and the deproteinized DNA samples were separated by 1% agarose gel electrophoresis. The average and standard deviation values of three independent experiments are shown in the bottom panel. (C) Human DMC1 (1 μM) was incubated with 32P-labeled ssDNA (3 μM), in the presence of RAD51 (lanes 3–6) or RAD51-GFP (lanes 7–10). The RAD51 and RAD51-GFP concentrations were 0.125 μM (lanes 3 and 7), 0.25 μM (lanes 4 and 8) and 0.5 μM (lanes 5, 6, 9 and 10). The reactions were initiated by the addition of superhelical dsDNA (100 μM) and continued at 37℃ for 10 min. Lane 1 is a negative control experiment without proteins. Lane 2 is a control experiment without RAD51 and RAD51-GFP, in the presence of DMC1. Lanes 6 and 10 are control experiments with RAD51 (0.5 μM) and RAD51-GFP (0.5 μM), respectively, in the absence of DMC1. The reactions were stopped by the addition of SDS and proteinase K, and the deproteinized DNA samples were separated by 1% agarose gel electrophoresis. The average and standard deviation values of three independent experiments are shown in the bottom panel.
A previous genetic study in A. thaliana found that RAD51-GFP complements the defective fertility of rad51/rad51 plants, indicating that it possesses the meiotic function (Da Ines et al., 2013). However, A. thaliana RAD51-GFP does not complement the mitotic DNA repair defect in the rad51/rad51 plants (Da Ines et al., 2013), consistent with earlier reports that human RAD51-GFP is also defective in mitotic DNA repair (Yu et al., 2003; Forget et al., 2007). These findings suggest that the GFP fused to the C terminus of RAD51 specifically interferes with the mitotic DNA repair function of RAD51. However, the mechanism underlying the mitosis-specific defect of RAD51-GFP has not yet been clarified. To answer this question, in the present study, we purified the rice and human RAD51-GFP fusion proteins and performed biochemical analyses.
We first found that the RAD51-GFP proteins efficiently bind to ssDNA and dsDNA (Fig. 1, B and C; Fig. 5, B and C). This finding is consistent with previous in vivo observations with A. thaliana, in which numerous RAD51-GFP foci were detected in nuclei of rad51/rad51 plants expressing exogenous RAD51-GFP, after DSB formation by gamma-irradiation (Da Ines et al., 2013). The foci are formed by RAD51 binding to ssDNA regions produced at the DSB sites (Tashiro et al., 2000; Kim et al., 2005). Importantly, the RAD51-GFP proteins are perfectly proficient in ssDNA-dependent ATP-hydrolyzing activity (Figs. 1D and 5D), suggesting that RAD51-GFP properly binds to ssDNA.
On the other hand, we found that RAD51-GFP complexed with ssDNA is significantly defective in secondary DNA binding, which is an essential step for homologous pairing (Figs. 2, 3 and 6). These biochemical data for the RAD51-GFP proteins suggest an explanation for how GFP fused to the C terminus of RAD51 inhibits mitotic DNA repair by homologous recombination. The crystal structures of S. cerevisae Rad51 filaments have been reported (Conway et al., 2004; Chen et al., 2010). In the model of the Rad51 filament structure, the GFPs fused to the C termini are located on the outside of the filament, and may restrict the access of the DNA molecule to the secondary DNA-binding sites (Fig. 7).

Model structure of the RAD51-GFP helical filament. (A) A model of the RAD51-GFP monomer. Structural data were obtained from the crystal structures of yeast Rad51 (gray; PDB ID: 3LDA) and EGFP (green; PDB ID: 2Y0G). Amino acid residues (R188, K361, K371) that are considered to be involved in the secondary DNA-binding site (Kurumizaka et al., 1999; Cloud et al., 2012) are colored magenta. The N-terminal domain of Rad51 is colored red. (B) A model of the RAD51-GFP helical filament. The black line represents the helical axis of the Rad51 filament. The dotted lines represent the Pro-Val-Ala-Thr linker peptide.
The secondary DNA-binding site was mapped by mutational analyses with the bacterial RAD51 homologue RecA (Kurumizaka et al., 1996, 1999). The corresponding Arg188, Lys361 and Lys371 residues of S. cerevisiae Rad51 are involved in the secondary DNA-binding site (Cloud et al., 2012). These residues are located on the solvent-accessible surface near the helical axis of the Rad51 filament (Fig. 7, magenta). In addition, the N-terminal domain of RAD51 also reportedly binds to DNA, and may function in secondary DNA binding (Aihara et al., 1999; Yu et al., 2001; Galkin et al., 2006). The RAD51 N-terminal domain may not constitute the secondary DNA-binding site, but could function to introduce the second DNA molecule into the secondary DNA-binding sites within the RAD51-ssDNA filament (Aihara et al., 1999; Yu et al., 2001). The C-terminally fused GFP portion is not located near the N-terminal domain of Rad51 (Fig. 7A). However, the GFPs may interfere with DNA binding by adjacent RAD51 N-terminal domains in the RAD51-GFP filament (Fig. 7B).
In contrast to its role in mitotic DNA repair, RAD51 may not function as a catalytic protein for homologous pairing in meiotic homologous recombination, but is suggested to participate as an accessory factor for stimulating DMC1 (Cloud et al., 2012). We found that RAD51-GFP retains the stimulatory activity for DMC1-mediated homologous pairing (Figs. 4C and 6C). This is consistent with previous genetic results with A. thaliana, in which RAD51-GFP fully complements the meiotic defects in rad51/rad51 plants (Da Ines et al., 2013). These results agree well with the idea that RAD51 possesses two distinct functions, homologous pairing and DMC1 stimulation, and that GFP fused to the C terminus of RAD51 specifically inhibits its homologous pairing activity by interfering with secondary DNA binding.
The distinct mitotic and meiotic RAD51 functions were originally detected in yeast genetic and biochemical analyses (Cloud et al., 2012). Our biochemical results with the rice and human RAD51-GFP proteins are quite compatible with a previous genetic study involving a plant, A. thaliana (Da Ines et al., 2013), and with complementation experiments involving RAD51–/– chicken DT40 cells or RAD51-knockdown human cells producing human RAD51-GFP (Yu et al., 2003; Forget et al., 2007). These findings suggest that GFP fused to the RAD51 C terminus commonly inhibits RAD51-mediated homologous pairing, but not its DMC1 stimulating activity, in yeast, plant and mammalian systems. Therefore, the RAD51-GFP protein should be an excellent tool to study the repair process of non-programmed DSB lesions that are potentially harmful for meiotic cell progression.
This work was supported in part by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Number 25250023 [to H. K.] and by MEXT KAKENHI Grant Number 25116002 [to H. K.]. H. K. was also supported by the Waseda Research Institute for Science and Engineering and Waseda University. W. K. and S. M. were supported by Research Fellowships of JSPS for Young Scientists.