2016 年 91 巻 6 号 p. 293-299
2016 年 91 巻 6 号 p. 293-299
Natural accessions are used for studying intraspecies genetic variation in the model plant Arabidopsis thaliana in order to address fundamental questions of evolution. Transposable elements are responsible for a wide range of mutations and play significant roles in shaping a genome over evolutionary time. In the present study, we aimed to characterize ONSEN, a heat-activated long terminal repeat (LTR) retrotransposon, in natural A. thaliana accessions. Southern blot analysis demonstrated that ONSEN was present in all the studied accessions, but the copy number was diverse. Olympia-1 contained a single ONSEN copy, located in the centromere of Chromosome 3. A premature stop codon in Olympia-1 ONSEN presumably abolishes integrase activity, which in turn presumably renders the retrotransposon non-functional. Hybridization of Col-0 with Olympia-1 showed that several ONSEN copies in Col-0 were activated by heat stress and maintained their transpositional activity in the progeny.
Arabidopsis thaliana has been used as a model organism in plant biology because of its relatively small genome and short life cycle. It is a predominantly self-pollinated plant and is widely distributed in the temperate zone (Maloof et al., 2001; Beck et al., 2008). More than 1,000 natural accessions have been isolated and characterized, revealing remarkable phenotypic variation in both morphological (e.g., leaf shape) and physiological (e.g., flowering time) traits (Ossowski et al., 2008; Atwell et al., 2010; Cao et al., 2011; Schneeberger et al., 2011). Natural genetic variation is considered to be partially influenced by changes in genome structure that are attributable to the abundance and distribution of transposable elements (TEs) (Madlung et al., 2012).
TEs cover a large portion of the genome in many plant species (e.g., more than 70% of the maize genome) and can affect gene expression when inserted near or within a gene-coding region (Sanmiguel and Bennetzen, 1998; Makarevitch et al., 2015). Previous studies have shown that TEs are regulated by epigenetic changes (DNA methylation or histone modification) and activated by environmental stress conditions (Chandler and Walbot, 1986; Bennetzen, 1987; Hirochika, 1993; Grandbastien et al., 1997; Scortecci et al., 1997; Steward et al., 2000; Hashida et al., 2003; Henderson and Jacobsen, 2007; Hirayama et al., 2009; Lisch, 2009; Zeller et al., 2009).
ONSEN is a heat-activated Ty1 / copia-like retrotransposon in A. thaliana (Pecinka et al., 2010; Tittel-Elmer et al., 2010; Ito et al., 2011). In addition, ONSEN-related copies have been found in most species of the family Brassicaceae (Ito et al., 2013). Eight full-length ONSEN copies in the reference accession Columbia-0 (Col-0) are regulated by heat-shock factors (HSFs) that recognize heat-shock elements (HSEs) in heat-activated genes (Cavrak et al., 2014). Of 21 HSFs in A. thaliana, HsfA2 regulates the activation of ONSEN, which contains HSEs in the promoter regions of its long terminal repeats (LTRs) (Cavrak et al., 2014). Three of the eight copies contain identical LTRs, indicating their relatively recent transposition. An extrachromosomal DNA is synthesized from an expressed ONSEN copy following activation by heat stress (Ito et al., 2011). In Col-0, extrachromosomal DNAs are derived from six of the eight ONSEN copies (Cavrak et al., 2014), but their transpositional activity remains unknown.
A mutant deficient in small interfering RNA (siRNA) biogenesis showed higher transcript levels of ONSEN, while transposition was detected in the progeny of a heat-stressed mutant, indicating that the activation of ONSEN is controlled by siRNA-mediated epigenetic regulation (Ito et al., 2011). However, transcriptional activation and mobility of the eight Col-0 ONSENs in a mutant background remain unclear. In the present study, we found that the accession Olympia-1 contains a single copy of ONSEN. Segregation of the Col-0 copies by crossing the siRNA mutant with Olympia-1 allowed us to characterize the transpositional activity of ONSEN copies subjected to heat stress.
The set of 96 accessions (CS22660) used for Southern blot analysis was previously described by Nordborg et al. (2005). Four natural accessions, Olympia-1 (CS75905), Faner-1 (CS75670), Tanz-1 (CS75924) and Tanz-2 (CS75925), were added for Southern blotting. The nrpd1a-3 mutant (Herr et al., 2005) was crossed with Olympia-1 for our hybrid analysis. The plants were grown on Murashige and Skoog (MS) plates at 21 ℃ under continuous light conditions.
Southern blot analysisGenomic DNA was isolated using the Nucleon PhytoPure DNA extraction kit (GE Healthcare Life Science, Chicago, IL, USA). Southern blotting was performed as described by Miura et al. (2004). Hybridization signals were detected using a radiolabeled ONSEN-specific probe (Supplementary Table S1), generated using the Megaprime DNA Labeling System (GE Healthcare Life Science). Genomic DNA of Col-0 was used as a template for the probe.
Heat stress treatmentSeven-day-old seedlings grown on MS plates were subjected to a temperature shift from 21 ℃ to 37 ℃ for 24 h. Two days after this heat treatment, the seedlings were transplanted to two-inch pots with expanded vermiculite and allowed to grow at 21 ℃.
Real-time PCRTotal RNA was extracted from whole seedlings grown on MS medium using TRI Reagent (Sigma Aldrich, St. Louis, MO, USA), according to the manufacturer’s instructions. Approximately 3–5 μg of total RNA was treated with RQ1 RNase-free DNase (Promega, Madison, WI, USA) and reverse-transcribed using the ReverTraAce qPCR RT Kit (Toyobo, Osaka, Japan) with an oligo(dT) primer. To quantify the amount of ONSEN DNA, genomic DNA was extracted from leaves using the DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA), according to the manufacturer’s instructions. Real-time PCR was performed using the Applied Biosystems 7300 Real Time PCR System with Thunderbird SYBR qPCR Mix (Toyobo). Three biological repetitions were performed, and standard deviation was calculated. DNA quantity was determined from a standard curve and normalized to the amount of 18S rDNA.
Sequence analysisA full-length ONSEN from the Olympia-1 genome was amplified by PCR. PCR primers (Supplementary Table S1) were designed to amplify AT3G32415 from the Col-0 genome (TAIR10 Whole genome). The first half of ONSEN was amplified with AT3G32415full-F and Olympia-seq-R primer pairs and the latter half was amplified using Maseq-1751 and AT3G32415full-R primers. The PCR fragments were sequenced after cloning into pGEM-T Easy Vector (Promega) (Supplementary Fig. S1).
Phylogenetic analysisPhylogenetic relationships were analyzed using the neighbor-joining method, and Jukes-Cantor distances were calculated from full-length ONSEN sequences. Indel sites were treated with the complete deletion option. All analyses were performed with MEGA 6.0 (Tamura et al., 2013). Bootstrap probabilities were estimated by 500 replications.
To identify the genomic copy number diversity of ONSEN, we conducted a Southern blot analysis with an ONSEN probe in 100 A. thaliana accessions. The results showed that ONSEN was present in all accessions, while the copy number varied among the accessions (Fig. 1, A and B). A single ONSEN copy was detected in Olympia-1 (Fig. 1B).
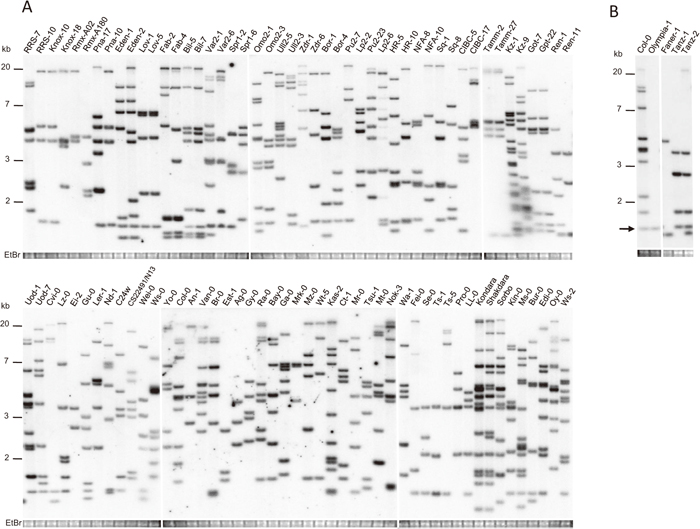
Southern blots of ONSEN copies in 96 Arabidopsis thaliana accessions (A) and four additional natural A. thaliana accessions (Olympia-1, Faner-1, Tanz-1 and Tanz-2) with the reference accession Columbia-0 (Col-0) (B). Genomic DNA was digested with EcoRV and hybridized with an ONSEN-specific probe. The arrow in (B) indicates the conserved copy in Col-0 and Olympia-1. A gel stained with ethidium bromide (EtBr) is shown at the bottom of each panel as a loading control.
PCR was conducted using a common internal primer within ONSEN (8copies-REVERSE) and a second primer in the flanking region that was specific for each copy of ONSEN in Col-0. The single ONSEN copy in Olympia-1 was conserved with the same flanking sequence as in Col-0 AT3G32415 (Fig. 2A). Its sequence was 99.6% identical to AT3G32415 (Supplementary Fig. S1) and phylogenetically closer to it than to other copies in Col-0 (Fig. 2B). Two of the predicted open reading frames of AT3G32415 were non-functional owing to the presence of stop codons within the gene-coding regions (Fig. 2C). The ONSEN copy in Olympia-1 also contained a stop codon in the same position within the integrase gene-coding region in Col-0 AT3G32415, indicating its transpositional deficiency (Fig. 2C). The two LTRs located at both ends of ONSEN in Olympia-1 were 98% identical to those in Col-0 AT3G32415, indicating that this copy is relatively old, because young copies contain 100% identical LTRs such as AT1G11265, AT3G61330 and AT5G13205 in Col-0 (Ito et al., 2011).

Sequence analysis of ONSEN copies in A. thaliana Olympia-1 and Col-0. (A) PCR analysis for detecting the conserved copy of ONSEN in Col-0 (C) and Olympia-1 (O). (B) Neighbor-joining tree of ONSEN copies in Olympia-1 and Col-0. The single copy in Olympia-1 and the eight copies in Col-0 are labeled in Supplementary Fig. S1. Bootstrap probabilities are shown near the branches. A scale bar indicating genetic distance (JC distance) is shown at the bottom left side of the tree. (C) The structure of Col-0 AT3G32415 and the ONSEN copy in Olympia-1. LTR: Long terminal repeat; Gag: Capsid protein; INT: Integrase; RT: Reverse transcriptase; RH: RNase H. Gray arrows mark open reading frames larger than 500 bp. Stop codons were found at the positions 1121 and 2714 bp. The region used as a probe for Southern blots is shown at the bottom.
To analyze heat activation of ONSEN in Olympia-1, we compared the sequences of HSEs in Olympia-1 and Col-0. HSEs were conserved in the eight ONSEN copies in Col-0 and the single copy in Olympia-1 (Fig. 3A). Heat activation of ONSEN was observed in Olympia-1; however, the transcript level of the single ONSEN copy in Olympia-1 was lower than that of the ONSENs in Col-0, suggesting that the former was not fully activated or transcribed by heat stress (Fig. 3B). Additionally, the transcript level of two heat-responsive genes, HsfA2 and HSP90.1, in Olympia-1 was up-regulated by heat stress, suggesting that HsfA2-mediated transcriptional activation can function in Olympia-1 (Fig. 3B).

Heat activation of ONSEN. (A) DNA alignment of the 81–156 bp section of LTRs in A. thaliana Olympia-1 and Col-0. Conserved heat shock elements (HSEs) (nTTCnnGAAnnTTCn or nGAAnnTTCnnGAAn) and possible HSEs (nGAAnnTTCor nTTCnnGAA) are highlighted. (B) Relative transcript levels of ONSEN, HsfA2 and Hsp90.1 in Olympia-1 and Col-0. Error bars represent the mean ± standard deviation (n = 3). Values are relative to heat-stressed Col-0. NS, non-stress; HS, heat stress. Relative transcript level of ONSEN was normalized by the copy number (Col-0:Olympia-1 = 8:1). The relative transcript levels of ONSEN, HsfA2 and Hsp90.1 in Col-0 were compared with the respective levels in Olympia-1 using a t-test. An asterisk indicates a significant difference, P < 0.05.
To analyze the mobility of each ONSEN copy in Col-0, we crossed Olympia-1 with Col-0 (nrpd1a-3). The F2 generation was genotyped for nrpd1a-3 mutations, and the ONSEN copies present in F2 genotypes were identified by PCR (Supplementary Table S1). nrpd1a-3-homozygous plants were heat-stressed, and their progeny were analyzed by Southern blotting. A new ONSEN copy was detected in an nrpd1a-3-homozygous Col-0/Olympia-1 progeny line that contained the ONSEN copies AT1G11265, AT1G48710, AT3G61330 and AT5G13205 (Fig. 4, A–C); however, no new ONSEN copies were detected in the progeny lines that contained only AT1G58140 and AT3G32415 (Fig. 4D). The results suggested that AT3G61330, AT5G13205, AT1G11265 and/or AT1G48710, but not AT1G58140 or AT3G32415, have transposition activity (Fig. 4B).

Transpositional and transcriptional activation of progeny lines derived from heat-stressed nrpd1-homozygous F2 Col-0/Olympia-1 plants. AT3G32415 and the single copy of ONSEN derived from Olympia-1 are indistinguishable and are named AT3G32415. Arrowheads show new ONSEN copies detected by Southern blotting in progeny lines that contained AT1G58140, AT3G32415 and AT3G61330 (A), AT1G11265, AT1G48710, AT1G58140 and AT3G32415 (B), or AT1G58140, AT3G32415 and AT5G13205 (C). No new ONSEN copies were detected in progeny lines that contained AT1G58140 and AT3G32415 (D). Corresponding bands of each ONSEN copy are indicated on the left side of (A). The first lane in each panel is the Col-0 parent line. A gel stained with ethidium bromide (EtBr) is shown as a loading control at the bottom of each panel. The upper of the two bands marked as AT1G48710 in (A) represents cross-hybridization of the probe with a genomic Col-0 DNA fragment that contains a region similar in sequence to ONSEN.
The copy number of some TEs differs among natural A. thaliana variants (Miura et al., 2004). Southern blot analysis showed that ONSEN was present in all the studied accessions, but the copy number in the genome varied among the accessions. The low frequency of each locus indicated that most copies cannot increase their frequency after transposition and are degraded. ONSEN was highly activated in the nrpd1a-3 mutant, and transgenerational transposition was observed in the progeny (Ito et al., 2011). The copy number of ONSEN in the analyzed accessions was lower than that in the nrpd1a-3 mutant subjected to heat stress, suggesting that somatic transposition was tightly controlled by small RNA-mediated epigenetic regulation and did not pass into the progeny, although transcriptional activation was induced by heat stress.
The expression level of ONSEN in Olympia-1 was approximately 16% of that in Col-0. In Olympia-1, ONSEN is located in the centromere of Chromosome 3, in which constitutive heterochromatin suppresses gene expression, indicating that the expression of ONSEN is influenced by a positional effect. Previously, we reported that the ONSEN copy located in the centromere was highly conserved in 53 out of 95 studied A. thaliana accessions; however, it did not affect the gene expression of the host plant and escaped from natural selection (Ito et al., 2013). Therefore, it would be useful to analyze the transcript level of each ONSEN copy in Col-0, although distinguishing between their transcripts is difficult.
Multiple copies of ONSEN are found in most A. thaliana accessions. To identify the ONSEN copy that was autonomous in Col-0, we crossed Col-0 with Olympia-1, which contains only a single copy of ONSEN. Since transposition could be induced in a mutant that was deficient in small RNA-mediated regulation, we crossed the nrpd1a-3 mutant (Col-0 background) with Olympia-1 to segregate copies of ONSEN in Col-0. Transposition of ONSEN was observed in several nrpd1-homozygous Col-0/Olympia-1 progeny lines that contained segregated ONSEN copies derived from Col-0. Three copies in Col-0 (AT1G11265, AT3G61330 and AT5G13205) contained identical LTRs, indicating their recent transposition. The transposition of multiple ONSEN copies was consistent with a previous study, which revealed that the integrase-coding region of six ONSEN copies in Col-0 is conserved, suggesting that they are potentially transposable, although their transposition frequency is probably regulated epigenetically (Cavrak et al., 2014).
In the present study, the frequency of transgenerational transpositions of ONSEN derived from Col-0 in the Col-0/Olympia-1 hybrid was variable (5–16% with one active copy, Fig. 4), indicating that transposition occurred with different frequencies among the individual copies. It has been reported that epigenetic modifications differ among A. thaliana accessions (Becker et al., 2011; Schmitz et al., 2013); therefore, it is worth considering that the transposition of ONSEN might be regulated by a hybrid-specific epigenetic modification. Genome-wide epigenetic information about Col-0 and Olympia-1 might allow us to obtain further detailed data on the underlying regulatory mechanisms.
We found that ONSEN was present in natural A. thaliana accessions, although the copy number was different among their genomes. Previous studies have reported an increase and decrease in the copy number of TEs in A. thaliana, as well as the selection of smaller genomes and the recent reduction of transposition, possibly owing to the transition to selfing (Hu et al., 2011; de la Chaux et al., 2012; Shimizu and Tsuchimatsu, 2015). ONSEN is regulated mainly by temperature, and the accumulation of multiple copies may have a deleterious effect on the host plants by destroying their gene function. Thus, the transposition–degradation cycle may determine the current ONSEN organization in natural A. thaliana accessions.
This study was supported by a Grant-in-Aid for Regional R&D Proposal-Based Program from the Northern Advancement Center for Science & Technology of Hokkaido, Japan, the NIG Collaborative Research Program (2015-A1 18), JST-PRESTO, a Grant-in-Aid for JSPS Fellows (14J02452), a Grant-in-Aid for Scientific Research on Innovative Areas (JP15H05960), and a Grant-in-Aid for Scientific Research in Innovative Areas (2511970103).