2023 年 98 巻 5 号 p. 283-286
2023 年 98 巻 5 号 p. 283-286
Duplicated genes show various degrees of functional diversification in plants. We previously identified 1,052 pairs of high diversified duplicates (HDDs) and 600 pairs of low diversified duplicates (LDDs) in Arabidopsis thaliana. Single knock-down of HDDs induced abnormal phenotypic changes because the other gene copy could not compensate for the knock-down effect, while single knock-down of LDDs did not induce abnormal phenotypic changes because of functional compensation by the copy gene. Here, focusing on one pair each of HDDs and LDDs, we performed transcriptome analyses in single-knock-down transgenic plants. The numbers of differentially expressed genes in single-knock-down transgenic plants were not different between HDDs and LDDs. Thus, functional compensation inferred by transcriptomics was similar between HDDs and LDDs. However, the trend of differentially expressed genes was similar in the pair of LDDs, while expression profiles were dissimilar in the pair of HDDs. This result indicates that a pair of LDDs tends to share similar functions but a pair of HDDs tends to have undergone functional divergence. Taking these findings together, as the reason for no phenotypic changes in single knock-down of LDDs but phenotypic changes in double knock-down of LDDs, we concluded that phenotypic changes of LDDs were induced by decreasing gene dosage.
There are many duplicate genes in plant genomes in comparison with other multicellular organisms (Moore and Purugganan, 2005). Some retained duplicates exhibit functional divergence, whereas others maintain redundant functions with their paralog. Functional divergence of duplicates can be inferred based on the phenotypes of knock-out mutants (Hanada et al., 2009, 2011). When single-knock-out mutants of a duplicate pair exhibit abnormal phenotypes, the mutations are not compensated for by the other gene copy. We have defined such duplicates as HDDs (Ezoe et al., 2021). When a double-knock-out mutant of both duplicates exhibits an abnormal phenotype in cases where an abnormal phenotype was not observed for single-knock-out mutants of either duplicate, the duplicates share redundant functions. These duplicates were defined as LDDs (Ezoe et al., 2021).
Pairs of HDDs tend to have higher divergence in their protein sequences and expression patterns compared with LDDs (Hanada et al., 2009). Based on the protein sequence divergence rate and the gene expression similarity rate, we developed a prediction method to infer HDDs and LDDs (Ezoe et al., 2021). To validate our classifications, we focused on two pairs of duplicates: one pair assigned as HDDs and the other pair assigned as LDDs. We then generated transgenic plants in which a single gene was knocked down (Ezoe et al., 2021). Single knock-down of HDDs is expected to induce abnormal phenotypic changes because there is little functional compensation. Conversely, single knock-down of LDDs induces few phenotypic changes because there is high functional compensation between the paralogs. Abnormal phenotypic changes were defined as a significant difference in either hypocotyl or root length at the seedling stage between transgenic and wild-type plants. As expected, single knock-down of HDDs induced abnormal phenotypic changes while single knock-down of LDDs had no significant effect on the phenotype of the transgenic plants. However, double knock-down of a pair of LDDs resulted in an abnormal phenotype. Thus, we showed that our assigned HDDs and LDDs tended to have low and high functional compensation, respectively.
However, our previous analyses did not directly evaluate the functional divergence in duplicate gene pairs. Here, instead of phenotypic changes, we performed transcriptome analyses to evaluate functional divergence in duplicate pairs. We focused on a tandemly duplicated pair (AT3G26270, AT3G26280) assigned as HDDs and another tandemly duplicated pair (AT1G18970, AT1G18980) assigned as LDDs. Both members of each duplicate pair had more than 90% amino acid sequence identity and 90% coverage in their alignments. In previous work, we generated AT3G26270i, AT3G26280i, AT1G18970i and AT1G18980i transgenic plants in which AT3G26270, AT3G26280, AT1G18970 and AT1G18980 expression, respectively, was knocked down via the overexpression of an artificial pre-miRNA sequence (Ezoe et al., 2021). A qRT-PCR analysis confirmed that the expression of target genes was significantly suppressed in the transgenic plants (Ezoe et al., 2021). Additionally, we prepared transgenic plants (vector control) containing only the vector as a negative control.
Seeds of transgenic Arabidopsis plants were sown on Murashige and Skoog plates and kept at 4 ℃ in darkness for at least two days to synchronize germination. Plants were grown at 22 ℃ under continuous light. Total RNA of the whole plant was extracted from two biological replicates of each transgenic plant at 14 days after sowing. Paired-end cDNA libraries (150 bp) were sequenced on an Illumina NovaSeq 6000 system. After trimming low-quality reads (phred score < 33, LEADING and TRAILING score < 20) with Trimmomatic version 0.39 software (Bolger et al., 2014), the qualified reads were mapped to 33,602 Arabidopsis representative genes (version 10, https://www.arabidopsis.org/) by Bowtie2 version 2.5.0 software (Langmead et al., 2019). For each sample, we obtained 37 to 54 million reads mapped to Arabidopsis genes (Table 1). We then calculated RPKM (reads per kilobase of exon per million mapped reads) as the expression intensity of each Arabidopsis gene. The trimmed read data generated in this study have been submitted to the Sequence Read Archive of the DNA Data Bank of Japan (https://www.ddbj.nig.ac.jp) under accession number DRA015520.
| Transgenic plant | Degree of divergence | No. of reads |
|---|---|---|
| pBST1_Rep1 | – (vector control) | 42,928,620 |
| pBST1_Rep2 | – (vector control) | 51,264,388 |
| AT3G26270i_Rep1 | High | 45,554,694 |
| AT3G26270i_Rep2 | High | 54,193,400 |
| AT3G26280i_Rep1 | High | 37,644,898 |
| AT3G26280i_Rep2 | High | 46,700,566 |
| AT1G18970i_Rep1 | Low | 53,207,282 |
| AT1G18970i_Rep2 | Low | 45,935,550 |
| AT1G18980i_Rep1 | Low | 43,123,842 |
| AT1G18980i_Rep2 | Low | 45,023,780 |
Using DESeq2 software (Love et al., 2014), we examined differentially expressed genes (DEGs) between the knock-down duplicate gene lines and vector control, and identified 135, 31, 64 and 178 DEGs in the AT3G26270i, AT3G26280i, AT1G18970i and AT1G18980i transgenic plants, respectively (false discovery rate < 0.05). Because pairs of LDDs tend to have high functional compensation in comparison with pairs of HDDs, we expected that the number of DEGs would be larger in the pair of HDDs (AT3G26270, AT3G26280) than in the pair of LDDs (AT1G18970, AT1G18980). However, the number of DEGs was not larger in HDDs than in LDDs, indicating that functional compensation inferred by DEGs does not occur more frequently in LDDs than in HDDs.
Furthermore, to address whether HDDs and LDDs tend to have low and high similarity in their DEGs, we examined the DEGs in the pair of HDDs (AT3G26270i vs. AT3G26280i) and the pair of LDDs (AT1G18970i vs. AT1G18980i). We identified 195 and 89 DEGs in the pairs of HDDs and LDDs, respectively (false discovery rate < 0.05). Thus, as expected, the pair of HDDs (AT3G26270 and AT3G26280) had more than twice as many DEGs as the pair of LDDs (AT1G18970 and AT1G18980). Transcriptome analysis therefore suggested that the pair of HDDs had lower similarity of DEGs than the pair of LDDs.
To evaluate the similarity of DEGs in the pairs of HDDs and LDDs, we focused on 156 DEGs in either AT3G26270i or AT3G26280i transgenic plants compared with the vector control as the pair of HDDs (false discovery rate < 0.05). After calculating fold changes between the two knock-down transgenic plants and the vector control, we compared the fold changes between AT3G26270i and AT3G26280i. The fold changes were not highly correlated (correlation coefficient R = 0.25, P-value = 0.02), indicating that the two transgenic plants had dissimilar expression profiles (Fig. 1A). We then identified 221 DEGs in either AT1G18970i or AT1G18980i transgenic plants compared with the vector control as the pair of LDDs (false discovery rate < 0.05). The fold changes (AT1G18970i/vector control vs. AT1G18980i/vector control) were positively correlated (R = 0.72, P-value = 1 × 10−35, Fig. 1B). These results indicated that the similarity of DEGs was lower in the pair of HDDs than in the pair of LDDs. Thus, as expected, our transcriptome data support the hypothesis that expression patterns tend to be more similar in the pair of LDDs than in the pair of HDDs.
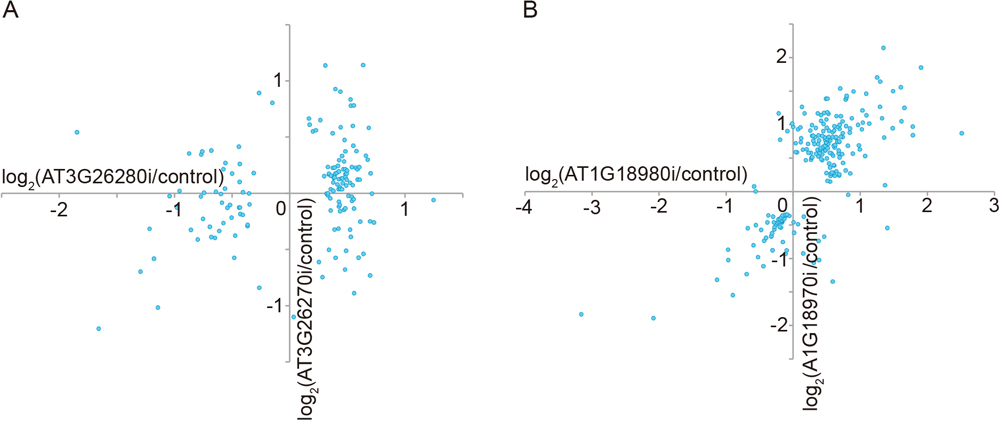
In the present study, transcriptome analysis showed that functional compensation in LDDs is not higher than that in HDDs with respect to DEGs. Nevertheless, knocking down of each LDD did not induce any phenotypic changes, while phenotypic changes were observed in the case of double knocking-down in the pair of LDDs (Ezoe et al., 2021). Because LDDs tend to regulate similar genes (Fig. 1B), double knocking-down of LDDs seems to induce stronger expression differences in the genes which were regulated by single knocking-down. Consequently, only the strong expression differences in double knocking-down of LDDs induced abnormal phenotypic change. That is, it is likely that phenotypic changes in LDDs are observed only when the level of gene expression is below a threshold. These results imply that the functions of genes that are regulated by LDDs are dependent on gene dosage.
HDDs tend to have lineage-specific functions related to biotic stress responses (low conservation and only in related species), whereas LDDs tend to be involved in essential pathways related to ubiquitous functions (high conservation in most organisms) (Ezoe et al., 2021). Genes associated with ubiquitous functions tend to be highly expressed and to have high gene dosage (Gout et al., 2010). Under natural environments, LDDs may be advantageous for increasing the gene dosage of ubiquitous functions. However, the single knocking-down of LDDs seems not to be detrimental under suitable laboratory conditions.
Funding: This study was supported by JSPS KAKENHI (JP23H04261, JP22H02675, JP20H03317 and JP18H02420 to K.H.; JP22K14870 to K.S.; JP23K13932 to A.E.), MEXT KAKENHI (JP21H05724, JP20H05905 and JP20H05906 to K.H.; JP22H05731 to K.S.), and the Asahi Glass Foundation (to K.H.). This study was partly supported by the Incentive Research Grant in RIKEN (to A.E.). A.E. was supported by the RIKEN Special Postdoctoral Researcher Program.
Conflicts of interest: The authors declare that there is no conflict of interest regarding the publication of this paper.
Ethics approval: This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent: None.
Authors’ contributions: Kousuke Hanada conceived and designed the research, and Tomoyuki Takeda and Akihiro Ezoe performed experimental analysis. All authors read and approved the final manuscript.
We thank Robbie Lewis, MSc, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.