2018 年 87 巻 4 号 p. 524-531
2018 年 87 巻 4 号 p. 524-531
For their exceptional beauty and great variety, waterlilies of the genus Nymphaea are popular ornamental plants worldwide. To improve their appealing traits, breeders worldwide have attempted intrasubgeneric and intersubgeneric cross-breeding and succeeded in producing new varieties. Molecular markers have become standard practice in the breeding process and for accurate variety identification. With increasing knowledge about transposable elements in eukaryotic genomes, retrotransposon-based markers have been developed for various plant species and were sometimes found to be more informative than conventional marker methods. Due to the lack of information about retrotransposons in waterlilies, this study aimed to confirm the presence and analyze the diversity of the Ty1-copia retrotransposons, which have been well studied and used to develop markers in many plants. 133 sequences of a conserved domain in the reverse transcriptase gene of Ty1-copia elements were isolated from 13 varieties of waterlilies and found to be homologous to either Ale, Angela, or TAR lineages. Sequences homologous to the Ale lineage were found to be abundant and diverse. Those homologous to the Angela lineage were found to be very conserved, but scarce. Those homologous to the TAR lineage were relatively conserved and were found in ample amounts. Furthermore, the results indicate that those homologous to the Angela or TAR lineages are more conserved to their counterpart in other plants than those homologous to the Ale lineage. From this study, 3 groups of Ty1-copia elements were found to be suitable candidates for development of retrotransposon-based markers for waterlilies in the future.
Waterlilies are aquatic plants in the genus Nymphaea, which consists of 45–50 species, and are found in habitats worldwide (Borsch et al., 2007). Nymphaea species are commonly categorized into 5 main subgenera: Anecphya, Brachyceras, Hydrocallis, Lotos, and Nymphaea, (Borsch et al., 2007; Conrad, 1905; Löhne et al., 2007). Although they are angiosperms, their evolutionary lineage has diverged from the lineage that gave rise to the majority of modern flowering plants, including monocots and dicots (Borsch et al., 2007; Soltis et al., 2008).
Due to their exceptional beauty, waterlilies are popular decorative plants worldwide. While other methods are available, cross-breeding between different varieties remains the primary procedure for producing new varieties of waterlilies with improved traits. The new varieties include hybrids from intrasubgeneric and intersubgeneric crosses (Doran et al., 2004; Li and Tao, 2008). In addition to morphological characterization, molecular marker techniques have become standard practice for precise variety identification. Techniques, such as DNA barcoding and PCR-RFLP, have been used to confirm parentage and identify new hybrids (Les et al., 2004; Songpanich and Hongtrakul, 2010). To further improve identification of increasing number of varieties, more molecular marker techniques should be developed for waterlilies.
Among different types of transposable elements, which are repetitive nucleotide sequences characterized by their capability to transpose to different genomic locations, Ty1-copia is an abundant and diverse superfamily of long terminal repeat (LTR) retrotransposons that has been extensively studied in plants (Schulman and Wicker, 2013). Ty1-copia retrotransposons are flanked at both ends by LTRs, which are structures essential for transposition and are conserved between LTR retrotransposons within the same family (Wicker et al., 2007). Various families of Ty1-copia elements have been identified in plant species (He et al., 2010; Weber et al., 2010; Wicker and Keller, 2007) and were found to make up a significant proportion of plant genomes (Hirakawa et al., 2012; Wicker et al., 2009).
Retrotransposon-based markers have been developed to take advantage of the abundance of LTR retrotransposons in plant genomes (Kalendar et al., 2011; Kumar et al., 1997). These markers are designed to detect polymorphisms in transposition patterns of these retrotransposons and have been found in some studies to be more informative than some conventional marker methods (Porceddu et al., 2002; Yu and Wise, 2000). Markers developed from nucleotide sequences of Ty1-copia retrotransposons have been developed and successfully used to study genetic diversity and identify cultivars and breeding lines of plants such as flax, grapevine, and wheat (D’Onofrio et al., 2010; Queen et al., 2004; Smýkal et al., 2011).
To the best of our knowledge, no study on Ty1-copia retrotransposons in Nymphaea species has been conducted, and no data essential for retrotransposon-based marker development is available. In this study, a polymerase chain reaction (PCR) procedure was applied to Nymphaea samples to isolate a conserved domain in reverse transcriptase (rt) gene of Ty1-copia retrotransposons present in their genomes. Preliminary analyses were conducted on these isolated fragments to confirm the presence of Ty1-copia retrotransposons in waterlilies and characterize their diversity. It is hoped that this study will provide information about the diversity of Ty1-copia retrotransposons in Nymphaea species that can be useful for marker development.
13 varieties of waterlilies in the genus Nymphaea and Victoria amazonica (family Nymphaeacea) were included in this study (Fig. 1). The Nymphaea samples include 2 varieties from the subgenus Anecphya, 5 from the subgenus Brachyceras, 1 from the subgenus Hydrocallis, 2 from the subgenus Lotos, and 3 from the subgenus Nymphaea. This collection covers all subgenera of the genus Nymphaea and represents the diversity of the genus. Genomic DNAs of these samples were extracted with a method modified from Doyle and Doyle’s protocol (1990) and diluted to a concentration suitable for PCR reactions.

Images of 13 samples from the genus Nymphaea and Victoria amazonica (N). Samples from the subgenus Anecphya included Nymphaea ‘Australian Giant’ (purple) (A), N. ‘Australian Giant’ (white) (B). Samples from the subgenus Brachyceras included N. ‘Chalong Kwan’ (N. ‘King of Siam’) (C), N. ‘Jongkolnee’ (N. siamensis) (D), N. ‘Nangkwak Fah’ (E), N. ‘Pech Chompoo’ (F), and N. ‘Suwanna’ (G). A sample from the subgenus Hydrocallis included N. prolifera (H). Samples from the subgenus Lotos included N. ‘Maeploi’ (I) and N. ‘Rojjana Ubol’ (J). Samples from the subgenus Nymphaea included N. ‘Mangkala Ubon’ (K), N. ‘Siam Nymph’ (L), and N. ‘Wanvisa’ (M).
PCR reactions using a degenerate primer pair designed by Flavell et al. (1992) were performed. Previous studies have shown that this primer pair is suitable for amplifying a conserved domain in the rt gene of Ty1-copia retrotransposons for phylogenetic analyses (Alipour et al., 2013; Kolano et al., 2013; Zedek et al., 2010). Each 50 μL reaction mixture contained 250 μM dNTPs, 5 mM MgCl2, 0.4 μM of each primer, 500 ng genomic DNA for each sample, and 2.5 Units of Taq polymerase recombinant (Invitrogen, CA, USA) in 1× PCR buffer. The PCR thermal profile consisted of an initial denaturation of 94°C for 5 min, 35 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 1 min, and a final elongation of 72°C for 10 min. Amplified DNA fragments of about 260 bps were purified and cloned into pGEM-T Easy vectors (Promega, WI, USA), and the vectors were transformed into the E. coli strain JM 109. For each sample, about 10 colonies with recombinant vectors were selected and cultured. Plasmids from these cultures were purified and sequenced with a BigDye Terminator v3.1 Cycle Sequencing Kit in an ABI Prism 3730xl Genetic Analyzer (Applied Biosystems, CA, USA).
Ty1-copia rt sequences screeningThe nucleotide sequences were checked against the National Center for Biotechnology Information’s (NCBI) database for homology to the rt gene in Ty1-copia elements and were translated into amino acid sequences. For amino acid sequences with the conserved domain disrupted by frameshift mutations, gaps were introduced to the nucleotide sequences to restore the correct open reading frames. Sequences that lacked homology to the rt genes or contained frameshift mutations that could not be confidently determined were excluded from subsequent steps in this study.
Classification of isolated retrotransposonsA widely accepted classification scheme by Wicker and Keller (2007) divides Ty1-copia retrotransposons into 6 major evolutionary lineages: Ale, Angela, Bianca, Ivana, Maximus, and TAR. 105 rt sequences from elements well classified in previous studies were gathered as references (Kolano et al., 2013; Lee et al., 2013; Llorens et al., 2009; Wicker and Keller, 2007) (Table S1). To classify the isolated rt sequences, each amino acid sequence was individually aligned to the reference sequences with ClustalW software for phylogenetic analyses. Each alignment was used to construct Neighbor-joining phylogenetic trees with 500 bootstrap replications and p-distance matrices with Molecular Evolutionary Genetic Analysis (MEGA) software (version 6) using the p-distance model and pairwise deletion treatment. The reference rt sequences most homologous to the isolated sequences were identified and used to determine the lineages that the isolated rt sequences could be in.
Phylogenetic analysis of Nymphaea Ty1-copia elementsTo determine the diversity of the isolated rt sequences, their amino acid sequences were aligned using ClustalW, and a Maximum Likelihood phylogenetic tree was constructed from this alignment with MEGA using the Jones-Taylor-Thornton model, pairwise deletion treatment, and 1000 bootstrap replications. In this tree, sequences were divided into groups according to the largest clusters with a bootstrap value of at least 75. Furthermore, p-distance matrices were computed between all sequences and sequences within each lineages and groups to find their average, maximum, and minimum p-distance value.
Comparison with Ty1-copia elements in other plantsDue to Nymphaea species’ evolutionary lineage, which has diverged from the majority of modern angiosperms, it is interesting to study the phylogenetic relationships between Ty1-copia retrotransposons from Nymphaea species and those from other plant species. A preliminary analysis was done by first checking the amino acid sequences of the isolated fragments against the NCBI database using the Standard Protein Basic Local Alignment Search Tool (BLASTp) with specificity to plant species. The isolated rt sequences and their most similar sequences from the database were aligned, and p-distances between them were computed. The average of these p-distances was also calculated for each group and lineage.
From 148 DNA fragments isolated, 15 sequences either had no homology to rt genes or contained unidentifiable frameshift mutations and were removed from the study. The remaining 133 sequences were further analyzed and can be found in Genbank database under accession numbers MG063487–MG063619. After the isolated rt were translated into amino acid sequences, it was found that 87 sequences (65.4%) had normal open reading frames while the other 46 (34.6%) contained premature stop codons and/or disrupted reading frames, which would most-likely interfere with gene function (Wicker et al., 2007) (Table 1).
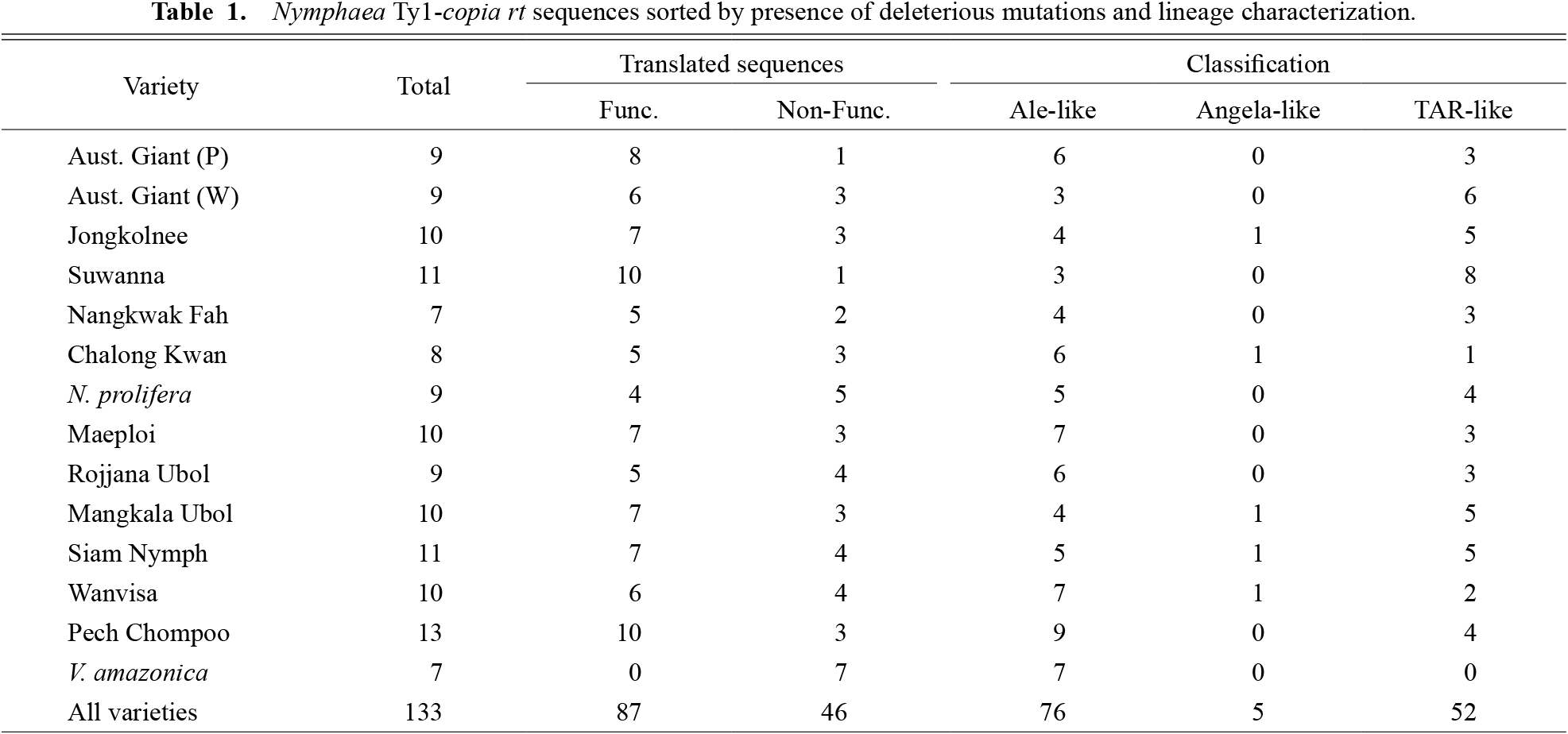
Nymphaea Ty1-copia rt sequences sorted by presence of deleterious mutations and lineage characterization.
From 133 isolated rt sequences, 70 sequences formed clusters with bootstrap values over 50 with at least one of the reference sequences. 18, 5, and 47 of these sequences formed clusters with references from the Ale, Angela, and TAR lineages, respectively. The reference sequences most homologous to the other 63 isolated sequences were determined with the p-distance values. 58 and 5 of these sequences showed the highest homology to references from the Ale and TAR lineages (Table S2). In total, 76, 5, and 52 isolated rt sequences were found to be the most similar to rt from the Ale, Angela, and TAR lineages, respectively (Table 1).
Although their most similar reference sequences were determined, many of the isolated sequences did not share significant homology with the reference sequences, despite prior confirmation of their homology to the rt gene in plant Ty1-copia retrotransposons by database checking. A major factor that could contribute to this heterogeneity between the isolated and reference sequences is the evolution of the waterlilies. The evolutionary lineage of the Nymphaea species has diverged early on from the lineage that gave rise to monocots and dicots, which the referenced elements came from (Borsch et al., 2007; Soltis et al., 2008). This divergence may have allowed some transposable elements in the two lineages to evolve independently and become significantly distinct. Because many did not have high homology to a lineage, the isolated elements most similar to Ale, Angela, and TAR lineages were instead dubbed as Ale-like, Angela-like, and TAR-like in the rest of the study.
So far, the rt sequences isolated from the samples have shown homology only to Ale, Angela, and TAR lineages. It could be speculated that only these three lineages are the only Ty1-copia retrotransposons present in Nymphaea species; however, the complete absence of elements from Ivana and Maximus lineages, which are abundant in dicots and monocots (Wicker and Keller, 2007), seems unlikely. Previous studies that used this isolation method in other plants also reported predominant isolation of elements closely related to Ale, Angela, and TAR lineages (Alipour et al., 2013; Kolano et al., 2013). This indicates a possible bias toward particular lineages based on the primers used, which was mentioned in other studies.
Phylogenetic relationship between Nymphaea Ty1-copia elementsIn the phylogenetic tree constructed from the isolated sequences (Fig. 2), 123 sequences were sorted into 18 groups according to the criteria mentioned previously while the other 10 sequences were not included in any group (Fig. S1). These groups included from 2 to 26 sequences coming from 2 to 11 samples, and their average p-distances ranged from 0.040 to 0.366 (Table 2). Thirteen groups were made up of Ale-like sequences, one was made up of Angela-like sequences, and four were made up of TAR-like sequences. The average p-distances for all sequences, Ale-like, Angela-like, and TAR-like sequences were 0.563, 0.542, 0.174, and 0.369, respectively.
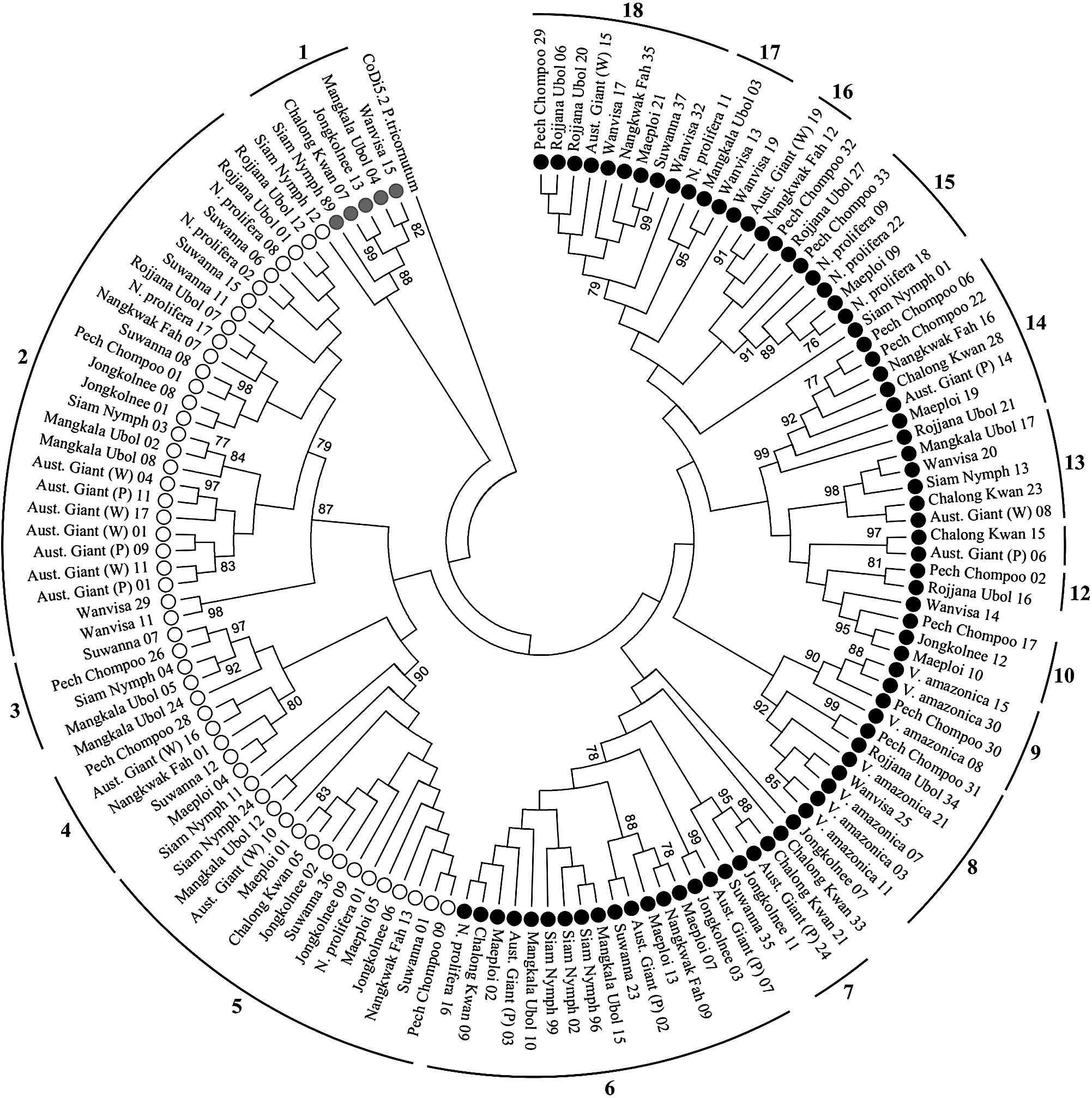
Maximum-Likelihood dendrogram of rt sequences isolated from the waterlily samples and rt sequence of CoDi5.2, a Ty1-copia retrotransposon from Phaeodactylum tricornutum used as an outgroup. Ale-like, Angela-like, and TAR-like sequences are marked with black, grey, and white circles, respectively.

Average, maximum, and minimum p-distance between sequences within each group, each lineage classification, and all sequences.
Some groups in the phylogenetic tree, such as groups 2, 5, and 6, consisted of a relatively large number of rt sequences from various samples, while others consisted of only a few sequences. These large groups included sequences from samples in all Nymphaea subgenera. This discrepancy in group size and source diversity indicates that some groups or families of Ty1-copia retrotransposons are more abundant and conserved throughout the genus Nymphaea than others. The abundance of transposon families generally depends on the rate of their proliferation and the rate of their removal from the genome (Schulman and Wicker, 2013).
It was observed that each type of isolated sequences had distinct characteristics. The Angela-like type consisted of few isolated sequences that made up only one group with the lowest average p-distance value. This result indicates that, while they may be scarce, Angela-like elements are notably conserved in Nymphaea species. The Ale-like type, on the other hand, was composed of the most numerous isolated sequences, which had the highest average p-distance and were divided into many small groups. This indicates that Ale-like retrotransposons are the most abundant and diverse and do not remain conserved between Nymphaea species. It has been noted that elements from the Ale lineage in plant genomes tend to be diverse and evolve rapidly (Du et al., 2010; Wicker and Keller, 2007). Lastly, while TAR-like sequences were not as numerous as Ale-like sequences, they made up a substantial proportion of total sequences and had lower average p-distance values than Ale-like sequences, and formed fewer groups than Ale-like sequences. This indicates that TAR-like elements are present in ample amounts and are relatively conserved in Nymphaea species. Although their complete diversity could not be represented with the method used, the results from this study showed some tendencies of major Ty1-copia lineages in the genus Nymphaea.
Comparison with Ty1-copia elements in other plantsWhen the isolated rt sequences were compared to the reference sequences for classification, it was found that Ale-like sequences generally shared lower homology to any of the references than Angela-like and TAR-like sequences did. The result from comparing these sequences to their most similar sequences from NCBI’s protein database also followed the same pattern. The calculated p-distance values between the isolated sequences and their most similar rt sequences from the database ranged from 0.169 to 0.583 (Table S3). These p-distances ranged from 0.301 to 0.583, 0.243 to 0.319, and 0.169 to 0.342 with average values of 0.392, 0.271, and 0.249 for Ale-like, Angela-like, and TAR-like sequences, respectively (Fig. 3).
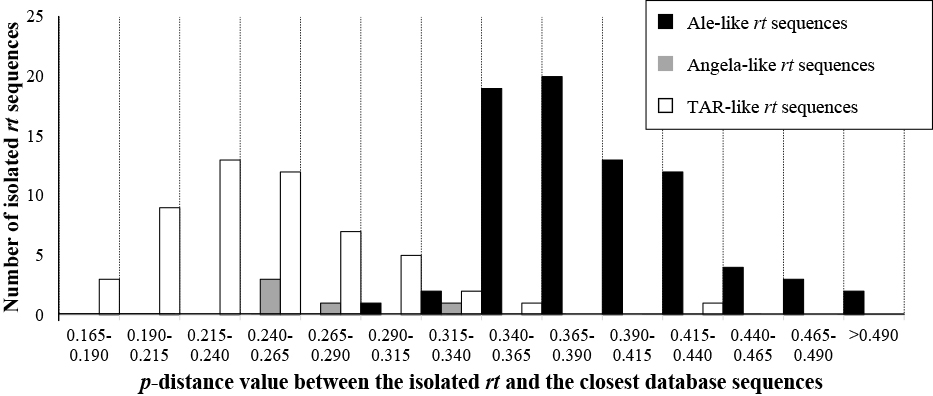
The number of isolated rt sequences within intervals of p-distance to their most homologous amino acid sequences from the NCBI database sorted by their lineage types. The height of the black, grey, and white bars corresponds to the number of Ale-like, Angela-like, and TAR-like sequences, respectively.
The overall p-distance values between the isolated sequences and their most similar sequences from the database were generally lower for Angela-like and TAR-like sequences than Ale-like sequences. This indicates that Ty1-copia elements in Nymphaea species most similar to the Angela and TAR lineages are more homologous to their counterparts in other plants than those most similar to the Ale lineage and that elements in the Angela and TAR lineages are more conserved between the evolutionary lineage of Nymphaea species and the lineage of monocots and dicots than those in the Ale lineage. Different degrees of conservation should be possible as some families of Ty1-copia elements and lineages of LTR-retrotransposons have been reported to be significantly conserved in a wide range of plant species (Du et al., 2010; Moisy et al., 2014; Smýkal et al., 2009).
Retrotransposon-based marker developmentMany retrotransposon-based markers, such as sequence-specific amplified polymorphism (SSAP) and inter-retrotransposon amplified polymorphism (IRAP), are PCR-based methods that utilize primers specific to conserved domains within LTR-retrotransposons to produce DNA fragments from multiple sites in genomes in which the retrotransposons are present (Kalendar et al., 2011). These fragments are then used as DNA fingerprints for identifying varieties and studying genetic diversity (D’Onofrio et al., 2010; Queen et al., 2004; Smýkal et al., 2011). For these PCR procedures to produce informative fingerprints, the primers should be designed to be specific to LTR retrotransposon families that are conserved between the studied samples so the techniques can be applied to all samples, and abundant in their genomes so numerous fragments are produced.
In addition to confirming their presence, this study provided information about the diversity of Ty1-copia retrotransposons in Nymphaea species that can contribute to the marker development process. The acquired results can be used to indicate the groups of Ty1-copia retrotransposons on which to focus according to the criteria mentioned above. Among the various groups, groups 2 and 5 are the most suitable for marker development because they comprised elements from most samples and were comparatively conserved (Fig. 2). Although group 6 was significantly less conserved than groups 2 and 5, attempts should also be made to develop markers from elements in this relatively large group (Table 2). While all LTR-retrotransposons can be used for marker development, elements from small groups may not be abundant enough in Nymphaea species for the markers to produce adequate fingerprints for studies.
The development of retrotransposon-based markers for waterlilies in the future will require nucleotide sequences of conserved domains, usually in LTRs, from families of LTR-retrotransposons in order to design primers, and different approaches using the isolated rt sequences can be used to acquire complete sequences of Ty1-copia retrotransposons from genomes of Nymphaea species which have not been reported. The rt fragments may be used as hybridization probes to screen genomic libraries for genomic fragments that contain the targeted retrotransposons as performed in Lilium longiflorum (Lee et al., 2013). Or the rt sequences may be used as the initiation site for genome walking to individually find nucleotide sequences of the desired transposons as done in Fragaria species (He et al., 2010). Conserved domains can then be determined by aligning complete sequences of elements in each Ty1-copia family and using them to design the primers. Retrotransposon-based markers developed in this manner have been successfully used to study genetic diversity of plant species (Lee et al., 2015; Melnikova et al., 2012).
The authors wish to thank “Pang U Bon”—Waterlily Garden, Thailand, Mr. Pairat Songpanich (waterlily hybridizer, Thailand) and Assist. Prof. Dr. N. Nopchai Chansilpa, Director of Waterlily Institute of Rajamangala Tawan-ok, Rajamangala University of Technology Tawan-ok, Chonburi, Thailand, for providing waterlily samples and some waterlily pictures used in this study.