2019 年 88 巻 4 号 p. 471-480
2019 年 88 巻 4 号 p. 471-480
In Japan, many turnip (Brassica rapa) cultivars are known as heirloom vegetables, especially in Shiga Prefecture, which is close to the old capital of Japan, Kyoto. Of these, ‘Omikabu’, an heirloom white turnip cultivar in Shiga Prefecture is referred to as the ancestor of the white turnip cultivars, ‘Shogoinkabu’, ‘Tennojikabu’, and ‘Yoriikabu’. Many “red turnip” cultivars that are red or purple-skinned varieties (and sometimes with colored flesh and petioles), also grow in Shiga Prefecture, and are mainly processed into pickled vegetables. However, their origins have not yet been fully verified. In this study, construction of neighbor-joining phylograms and population structure analyses were performed based on eight simple sequence repeat markers for white and red turnips, plus two non-turnip B. rapa vegetables (Chinese cabbage and mizuna). For the white turnip-related lines, a claim that ‘Shogoinkabu’, ‘Tennojikabu’, and ‘Yoriikabu’ are derived from ‘Omikabu’ could not be supported in this study because ‘Omikabu’ lines were separated from the above three cultivars in the phylogram. In contrast, an ‘Omikabu’ line, ‘Omikabura RU’, formed a cluster with ‘Jonansensuji mizuna’, suggesting a genetic relationship (a crossing in the past) between them. For red turnips, close placements of ‘Kisobenikabu’-‘Shinshukabu’, ‘Hinona’-‘Kitanoshokabu’, and ‘Biwakobenikabu’-‘Yurugikabu’ were found in the phylogram, each of which was in good agreement with the proposed cultivar’s origin. The data in this study provide useful information for understanding the genetic relationships among Japanese heirloom turnip cultivars.
Turnip (Brassica rapa) is cultivated worldwide as a vegetable and fodder. The main edible part is a combination of the swollen hypocotyl and root, although the leaves are sometimes eaten. In Japan, many turnip cultivars are known as heirloom vegetables, especially in Shiga Prefecture, which is close to the old capital of Japan, Kyoto. Some of the Shiga cultivars are thought to be the ancestors of turnip cultivars from other regions. For example, ‘Omikabu’, an heirloom turnip cultivar in Shiga is referred to as the ancestor of three turnip cultivars in Kyoto, Osaka, and Niigata Prefectures, ‘Shogoinkabu’, ‘Tennojikabu’, and ‘Yoriikabu’, respectively (Aoba, 1981; Kanda and Kadono, 1941c; Konoshima, 1993; Kyoto-fu Nokai, 1909; Otani, 1987; Shibutani, 1950). Many heirloom cultivars of “red turnip” varieties with red or purple pigmentation on the skin of the swollen hypocotyl and root (and sometimes on the flesh and petioles), also grow in Shiga Prefecture, and are mainly processed into pickled vegetables (Aoba, 1981; Kanda and Kadono, 1941a; Nagayoshi, 1959). Some of their ancestors have been proposed. For example, ‘Hinona’, a red turnip cultivar in Shiga Prefecture may be the ancestor of ‘Iyohikabu’ and ‘Matsuzaka akana’ in Ehime and Mie Prefectures, respectively (Konoshima, 1993; Osa and Osa, 2013). ‘Kitanoshokabu’ is a red turnip cultivar locally produced in Omihachiman, Shiga Prefecture, which seems to be a variety of ‘Hinona’ (Otani, 2004). ‘Hiruguchikabu’ and ‘Iriekabu’ in Shiga Prefecture could have been bred from ‘Koizumikabu’ in the same Prefecture (Kanda and Kadono, 1941b; Otani, 2004; Shibutani, 1950; Yoshizawa, 1987). ‘Shinshukabu’ in Shiga Prefecture is thought to be a variety of ‘Kisobenikabu’ that originated in Nagano Prefecture (Kanda and Kadono, 1941c; Otani, 1987). Many other ancestors have been proposed in literature and folklore. However, the ancestors have not yet been scientifically verified. Classification of heirloom turnip cultivars in Japan has been conducted based on morphological traits such as seed coat mucilage, leaf pubescence, plant shape, and bolting, etc. (Aoba, 1958, 1961, 1963; Ohi and Okada, 2000; Ohi and Sato, 2002; Shibutani and Okamura, 1954; Shibutani et al., 1952; Takahashi et al., 2016), by which Japanese turnips have been classified as Japanese, European and intermediate types.
Molecular markers are useful tools for genetic analyses such as classification, linkage mapping, and positional cloning of genetic loci. Genetic relationships of B. rapa vegetables including turnips have been examined using dominant molecular markers such as randomly amplified polymorphic DNA and amplified fragment length polymorphism markers (Fujimoto and Yamagishi, 1996; Pino Del Carpio et al., 2011; Takuno et al., 2007; Zhao et al., 2005). Simple sequence repeats (SSRs) consisting of 1–6 nucleotide repeat units are also used as molecular markers in many eukaryotic organisms because of their advantages such as abundance in eukaryotic genomes, high rates of polymorphism and stability, and relatively ease of detection of different alleles (see Vieira et al., 2016 for a review). Moreover, SSRs are codominant markers, capable of detecting heterozygotes, and this is a merit over dominant markers and informative for analyzing outcrossed species such as Brassica. Takahashi et al. (2016) classified turnip accessions in Japan and foreign countries based on SSR markers to overview the geographic origin of Asian turnips, but they did not produce a detailed classification of Japanese turnip cultivars.
In this study, we classified turnip cultivars in Shiga and other Prefectures based on SSR markers to verify their ancestors and relationships. The objective of this study was to obtain a close-up view of the genetic relationships among heirloom vegetables, using those in Shiga Prefecture as a representative case.
A total of 28 lines (eight white turnips, 18 red turnips, and two other B. rapa vegetables) were used in this study (Table 1). Ten to twenty (average 16.6) individuals per line were investigated for DNA analysis. Genomic DNA was extracted from fresh leaves using a DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA), or a crude DNA lysate was prepared from an approximately 5 mm2 leaf piece according to the method of Thomson and Henry (1995) and the instruction manual for KOD FX Neo (Toyobo, Osaka, Japan). Allele data on eight Chinese cabbage SSR markers (Suwabe et al., 2002, 2006), showing clear amplification and polymorphisms in our samples from preliminary experiments, were analyzed according to a previous study (Kubo et al., 2009, 2019). Data on a Chinese cabbage cultivar ‘Muso’ from the previous study (Kubo et al., 2019) were included for further analysis.

List of turnips and other Brassica rapa vegetables used in this study.
Numbers of alleles per locus (A), allelic richness (AR, a measure of the numbers of alleles independent of sample size) (Petit et al., 1998), observed (HO) and expected (HE) heterozygosities, and the fixation index (FIS) were calculated with GENEPOP 4.2 (Rousset, 2008) and FSTAT 2.9.3 (Goudet, 1995, 2001). Deviation of FIS from Hardy-Weinberg equilibrium (HWE) and linkage disequilibrium (LD) for each locus pair across all populations was tested with FSTAT 2.9.3. Analysis of molecular variance (AMOVA) was performed using Arlequin 3.5.2.2 (Excoffier and Lischer, 2010). Individual-based phylograms were constructed using Populations 1.2.32 (Langella, 2011) based on the neighbor-joining (NJ) method (Saitou and Nei, 1987) for white turnip-related lines and red turnips separately. We first constructed a combined version phylogram containing all the 28 lines, resulting in a highly confusing relationship with that of the structure analysis (data not shown). Therefore, we omitted the combined version in this study. The Chinese cabbage cultivar ‘Muso’ was used as a phylogram outgroup. Bootstrap analysis was performed with 1,000 replications using Populations 1.2.32.
Population structure analysisDetection of a hierarchical genetic population structure was performed with STRUCTURE 2.3.4 (Hubisz et al., 2009) with 50,000 burn-in steps and 1,000,000 Markov chain Monte Carlo steps after burn-in. A suitable number of subpopulations (K) was determined based on the ΔK values (Evanno et al., 2005) with STRUCTURE HARVESTER 0.6.94 (Earl and vonHoldt, 2012). Bar plots at determined K values were drawn with CLUMPAK 1.1 (Kopelman et al., 2015).
In the eight SSR markers, A and AR values ranged from 3 to 21 and from 2.758 to 7.780, respectively (Table S1). The average value of A in this study (8.63) was similar to that in previous reports on turnips (8.72) and the flower nabana (8.75) (Kubo et al., 2019; Takahashi et al., 2016). HE ranged from 0.0236 to 0.5959 (Table 2). The HE values were slightly lower than those in previous reports (Pino Del Carpio et al., 2011; Takahashi et al., 2016) presumably because local heirloom cultivars may have been maintained in a relatively small population. FIS values significantly deviated from HWE in only three turnip cultivars (Table 2, asterisks), so most of the given cultivars could be treated as randomly mating populations.

Genetic diversity of 28 B. rapa lines analyzed in this study.
Among the eight SSR loci, a pair of loci (BRMS-085 and BRMS-194) showed a significant LD (Table S1, asterisks). These loci were localized in close proximity (average map distance = 1.7 cM) on the linkage group (LG) A10 in our previous map (Inoue et al., 2015). The other pairs of loci did not show any LD, even though three of them (BRMS-158, BRMS-208, and BRMS-269) were mapped on the same LG, A3 (Table S1). Therefore, there were actually seven independent loci. The number of DNA loci used is important for population analysis. Although tens of SSR loci result in sufficient stability, population structures can only be inferred with six or more markers (e.g., Politov et al., 2015; Urrestarazu et al., 2015).
With the present SSR data set, AMOVA revealed that 49.08% and 50.92% of the variance occurred among and within populations, respectively (Table S2), suggesting significant (P < 0.001) genetic differences among cultivars. This was also supported by a high FST value (0.491) (Table S2). These results suggest that the cultivars investigated in this study were well differentiated. Because of mostly equal values of variance among and within cultivars, we made an individual-based phylogram to evaluate both inter- and intra-cultivar variations.
Overall relationships of white turnipsAn individual-based NJ phylogram was constructed for eight white turnips and ‘Jonansensuji mizuna’, using a Chinese cabbage cultivar ‘Muso’ as an outgroup (Fig. 1). Most individuals of the nine ingroup lines clearly clustered except for ‘Tennojikabu’. Individuals of this cultivar were separated to three locations in the phylogram. The resulting phylogram could be classified into three groups (I–III). Group I was composed of four ‘Tennojikabu’ individuals. Group II mostly included three ‘Omikabu’ lines and ‘Jonansensuji mizuna’. Group III was composed of the rest of the white turnip cultivars, including ‘Shogoinkabu’.
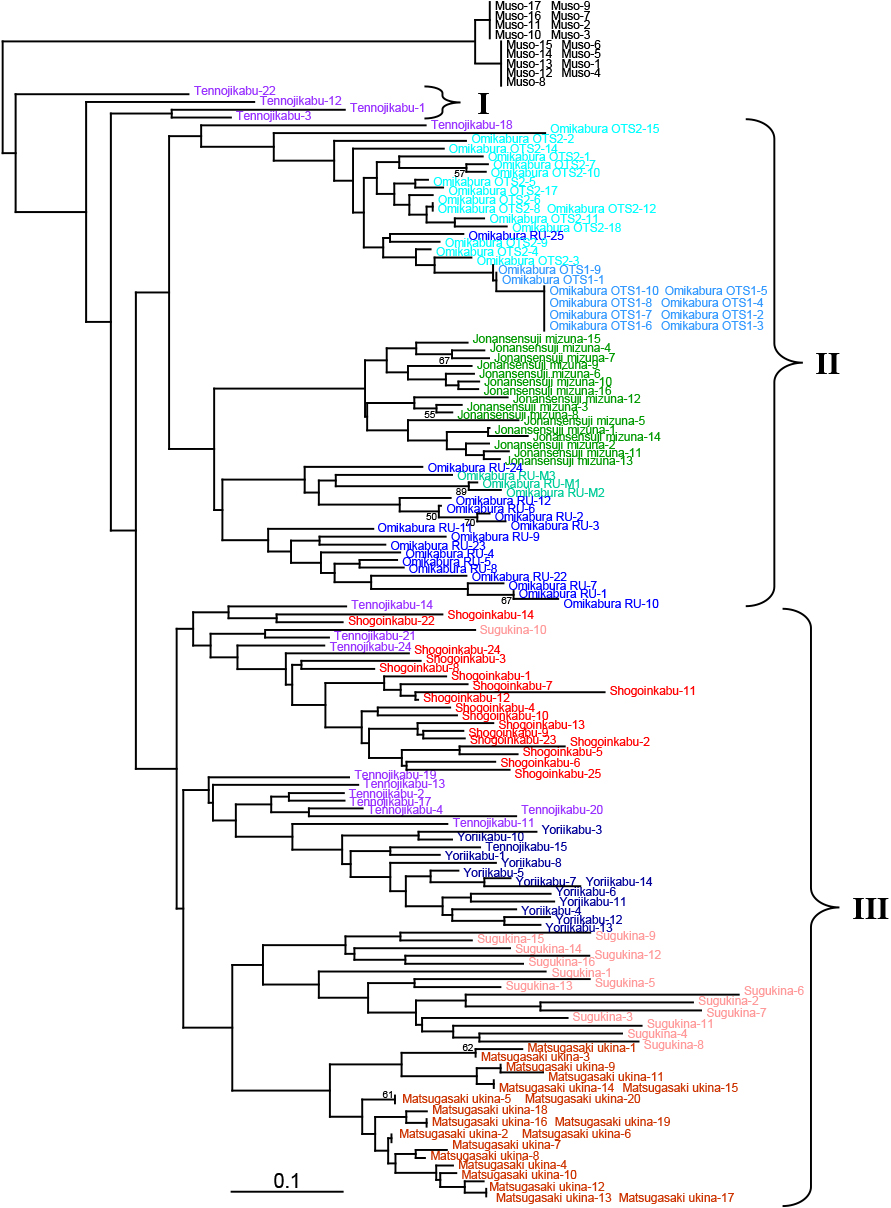
A neighbor-joining (NJ) phylogram of eight white turnips and two other B. rapa vegetables based on eight B. rapa SSR loci. Three potential groups are indicated with roman numbers (I–III). Individuals of each cultivar are differentially colored (see online article for color version of the figure). Numbers on nodes are bootstrap values from 1,000 replicates (≥ 50%). The scale bar indicates the genetic distance DA (Nei et al., 1983).
Three ‘Omikabu’ lines, ‘Omikabura RU’, ‘Omikabura OTS1’, and ‘Omikabura OTS2’, formed mostly independent groups based on each line (Fig. 1). Interestingly, a non-turnip B. rapa vegetable, ‘Jonansensuji mizuna’, was included in this group. Especially, ‘Omikabura RU’ (a line maintained in Ryukoku University) was placed in a neighbor group of ‘Jonansensuji mizuna’. In addition, individuals having serrated leaves and tens of branches that resemble mizuna occurred in the ‘Omikabura RU’ line at a 2.8% frequency (data not shown). Three such individuals (‘Omikabura RU’-M1 to ‘Omikabura RU’-M3) were included in the ‘Omikabura RU’ group (Fig. 1). These results suggest a relationship between these ‘Omikabu’ and mizuna lines. One reason for this may have been an outcrossing of mizuna during the maintenance of the current ‘Omikabu’ line, but another hypothesis has been provided by Aoba (1981): ‘Omikabu’ may be derived from a cross between turnip and mizuna although the timing of this cross is unknown. A relationship between the current ‘Omikabu’ and mizuna lines was also detected by population structure analysis (Figs. 2 and S1). ‘Omikabura RU’ shared the same cluster with ‘Jonansensuji mizuna’ at the highest appropriate number of subpopulations (Table S3, K = 5) (Fig. 2, red-purple color). The other two ‘Omikabu’ lines, ‘Omikabura OTS1’ and ‘Omikabura OTS2’, were separated from ‘Jonansensuji mizuna’ possibly due to having undergone different selection.

Hierarchical population structure of eight white turnip-related lines and two non-turnip B. rapa vegetables (‘Muso’ and ‘Jonansensuji mizuna’). The result from subpopulation (K) at 5 is shown. Populations are separated by vertical gray lines, and their names are listed below bar plots. Individuals are indicated with bar plots with colors corresponding to the sum of assignment probabilities to the clusters in each subpopulation (see online article for color version of the figure).
No ‘Shogoinkabu’ individual was included in any of the ‘Omikabu’ lines, both of which were placed in different groups (II and III). This result suggests a split of these two turnip cultivars. The split of ‘Omikabu’ and ‘Shogoinkabu’ was confirmed based on the genetic structure. These two cultivars were mainly assigned to different clusters in any of three subpopulations (K = 3–5) (Fig. S1), although ‘Omikabura RU’ slightly shared some clusters (Fig. S1, blue color). A great deal of the literature indicates a relationship between ‘Omikabu’ and ‘Shogoinkabu’ (Aoba, 1981; Kanda and Kadono, 1941c; Konoshima, 1993; Kyoto-fu Nokai, 1909; Otani, 1987; Shibutani, 1950). In older literature, “Seikei-Zusetsu” (So and Shirao, 1804) and “Honzo-Zufu” (Iwasaki, 1828), ‘Omikabu’ was drawn as a turnip with a spherical shape, whereas the current ‘Omikabu’ has an extremely ellipsoidal shape. ‘Tojikabu’, classified as an ‘Omikabu’-related cultivar (Aoba, 1981; Kumazawa, 1959; Matsumura, 1988; Shibutani, 1961; Shibutani and Okamura, 1954), was also drawn as a spherical turnip in a painting (Yoshino, 2005). Based on this discrepancy, another “Omikabu” line with different morphology from the current line may have existed in the past. An extinct turnip cultivar with a spherical shape in Shiga Prefecture, ‘Hyozukabu’, may have been confused with the current ‘Omikabu’ because both ‘Hyozukabu’ and ‘Omikabu’ used to be cultivated in the same district (an ancient province, Omi), and both were known as “Omikabu” (Kojima and Takamasa, 2009; Konoshima, 1993; Kubo, 1997).
Relationships among miscellaneous white turnips‘Tennojikabu’ and ‘Yoriikabu’ are also proposed to be ‘Omikabu’-derived cultivars (Otani, 1987; Shibutani, 1961). Such a proposal was not supported in the present phylogram as in the case of ‘Shogoinkabu’, in which no ‘Omikabu’ individual was grouped with the ‘Tennojikabu’ or ‘Yoriikabu’ lines. However, ‘Tennojikabu’ may have some genetic relationship with ‘Shogoinkabu’ and ‘Yoriikabu’ because some ‘Tennojikabu’ individuals were included in clades of these two cultivars in the phylogram. Individuals of ‘Tennojikabu’ formed three clusters in the present phylogram (Fig. 1, purple color). Our preliminary analysis using ‘Tennojikabu’ from another seed company also resulted in the formation of a few clusters (data not shown). Therefore, the ‘Tennojikabu’ cultivar we used could contain a relatively high number of variations, presumably occurring during the course of its establishment and maintenance.
‘Sugukina’ and ‘Matsugasaki ukina’ are cultivars in Kyoto Prefecture, and their origins are unclear. Takashima and Narita (1971) reported the origin of the current ‘Sugukina’ as being derived from a cross between an ancestral ‘Sugukina’ population that was already extinct and ‘Shogoinkabu’. In the present phylogram, ‘Matsugasaki ukina’, ‘Shogoinkabu’, and ‘Sugukina’ were included in the same group (III). This suggests a genetic relationship and/or a geographic connection among them as Kyoto turnips.
Relationships among red turnipsAnother individual-based NJ phylogram was constructed for 18 red turnips using a Chinese cabbage cultivar ‘Muso’ as an outgroup (Fig. 3). Most individuals of the 18 ingroup cultivars clustered except for ‘Yamakabu’. Individuals of this cultivar were scattered to three locations throughout the phylogram, forming groups with a few individuals. Among red turnips in Shiga Prefecture, ‘Shinshukabu’ was placed as the most basal turnip cultivar examined in this study and ‘Kisobenikabu’ was placed in a neighboring group. Their close placement was confirmed in an unrooted NJ phylogram (data not shown). ‘Kisobenikabu’ and ‘Shinshukabu’ are European types, whereas the rest of the red turnip cultivars investigated here are mostly the Japanese type (Aoba, 1958; Shibutani and Okamura, 1954). These results well supported their close relationship and the proposed relationship of ‘Shinshukabu’ as a variety of ‘Kisobenikabu’ (Kanda and Kadono, 1941c; Otani, 1987).

An NJ phylogram of 18 red turnips and one other B. rapa vegetable based on eight B. rapa SSR loci. Other legends refer to those in Figure 1. See online article for color version of the figure.
Several cultivars are suspected to be derived from ‘Hinona’. Relationships among ‘Kitanoshokabu’ and ‘Matsuzaka akana’ with ‘Hinona’ were suggested in this study based on the fact that they were included in the same cluster in the lower part of the phylogram (Fig. 3). The slightly different position of the current ‘Matsuzaka akana’ from ‘Hinona’ suggests genetic drift from the original population by re-selection (Osa and Osa, 2013). A well known proposed relationship that ‘Iyohikabu’ was bred from ‘Hinona’ was not suggested in this study because they were placed in different groups. Similarly, no relationship was suggested between ‘Iyohikabu’ and ‘Yurugikabu’. No evidence supporting relationships among ‘Koizumikabu’, ‘Hiruguchikabu’ and ‘Iriekabu’ was obtained in this study. According to the literature, the latter two cultivars were bred from ‘Koizumikabu’ ca. 400 and 150 years ago, respectively (Kanda and Kadono, 1941b; Otani, 2004; Shibutani, 1950; Yoshizawa, 1987). Each of the three cultivars was distantly placed in the phylogram (Fig. 3). We presently have no idea to explain such a discrepancy.
‘Iriekabu’, ‘Maizurukabu’, and ‘Tsudakabu’ from Shiga, Kyoto, and Shimane Prefectures, respectively, formed a cluster, but the reason is unclear. Finally, ‘Yurugikabu’ and ‘Biwakobenikabu’ also clustered together, implying some yet-to-be-defined relationship between them. ‘Biwakobenikabu’ was selected from a ‘Yurugikabu’ line with superior red pigmentation (Ota Seed Co., Ltd., Japan; personal communication). The close relationships described above were also confirmed by the genetic structure, in which close cultivar groups shared the same subpopulations at the highest appropriate number of subpopulations (Table S4, K = 3) (Fig. 4, colors of bar plots) and even at any of three subpopulations (K = 3–5) (Fig. S2).

Hierarchical population structure of 18 red turnips and one non-turnip B. rapa vegetable (‘Muso’). The result from subpopulation (K) at 3 is shown. Other legends refer to those in Figure 2. See online article for color version of the figure.
Concerning skin color, cultivars with purple skins (Table 1) appeared to be linked to those with red skins. Such distributions did not contradict any relationships or pedigrees of the given cultivars, assuming that segregation of alleles occurred in the anthocyanin biosynthetic genes and associated trans-acting factors that are responsible for red and purple skin color (Chaves-Silva et al., 2018; Guo et al., 2014, 2015; and references therein). Different responsible genes may have independently occurred in different lines.
Effectiveness of genetic analysis with SSR markers to verify the origins of heirloom turnip cultivarsIn this study, we examined the genetic relationships among heirloom turnip cultivars, especially for cultivars in Shiga Prefecture. This region is located in almost the middle of Japan, near the old capital Kyoto, so it was an important traffic route connecting eastern and western provinces. Many crop varieties would have been brought and bred there. Although in our study a few terminal nodes were supported with relatively high bootstrap values, no large group was supported by a ≥ 50% bootstrap value. This could be because many of the lines were closely related and because some cultivars may be derived from intercrossing between distantly related lines. With the present SSR marker set, we obtained an average A value similar to that in other reports (Kubo et al., 2019; Takahashi et al., 2016). We successfully classified flowering nabana lines according to the parts used and cultivar origins with the same SSR marker set (Kubo et al., 2019). Therefore, we hope our data have further clarified the genetic relationships among heirloom turnip cultivars. Combined with further analyses using cultivars from other regions, the data in this study will provide useful information for understanding the genetic relationships among Japanese heirloom turnip cultivars.
We thank Shiga Prefecture Agricultural Technology Promotion Center, Agriculture, and Forestry Technology Department, Kyoto Prefectural Agriculture, Forestry, and Fisheries Technology Center, and Genetic Resources Center, NARO for providing turnip seeds. We are grateful to Mr. S. Osa, Dr. Y. Mimura, and Ms. H. Kasaoka for helpful comments and technical assistance, respectively. This work was partly supported by a grant from Research Institute for Food and Agriculture, Ryukoku University (2017–2018) to S.S.