2019 年 88 巻 4 号 p. 435-443
2019 年 88 巻 4 号 p. 435-443
The red coloration of the mango ‘Irwin’ skin is an important factor determining its value in the Japanese domestic luxury fruit market. In the present study, to investigate the molecular mechanism underlying anthocyanin biosynthesis of mango fruit skin, UFGT-like genes were isolated and the expression profile of anthocyanin-related genes was determined. Several UFGT-like genes were identified in transcriptome data of red ‘Irwin’ mango skin and two genes, MiUFGT1 and MiUFGT3, were considered to be involved in mango skin coloration. Deduced amino acid sequences of these genes exhibited high similarity to other plant UFGTs and contained the conserved PSPG box common to the glycosyltransferase family. The presence of a glutamine and a histidine residue at the C-terminus end of the PSPG box in MiUFGT1 and MiUFGT3, respectively, implied that MiUFGT1 and MiUFGT3 use glucose and galactose, respectively, as a sugar donor; however, the actual function and sugar donor preference of these enzymes remain to be elucidated. Expression analysis of anthocyanin-related genes during skin coloration suggested that MiCHS and MiANS, as well as MiUFGT1 and MiUFGT3, play important roles in the anthocyanin biosynthesis of mango fruit skin and that the expression of these genes is regulated by the MYB transcription factor, as reported in other plant species.
Mango (Mangifera indica L.) production in Japan has increased rapidly over the past three decades, reaching 3,800 tons in 2014, which was more than 100-times higher than in 1984 (Ministry of Agriculture, Forestry and Fisheries of Japan, 2018). With increasing production, mango has become a popular fruit in Japan and has been recognized as a luxury fruit for consumers. Japanese domestic mangoes are sold at a much higher price than imported mangoes, and some high-quality branded mangos are traded at very high prices (Nakakubo, 2009). In addition to taste, appearance is an important factor for obtaining high market value. To date, Japanese mango production has relied heavily on one cultivar ‘Irwin’, which is a Floridian cultivar with bright red skin. As complete red-coloration of the skin is a prerequisite for the luxury fruit market, growers focus on ensuring ‘Irwin’ fruits are consistently bright red.
Anthocyanins, the red pigments in mango skin, are the largest subclass of plant flavonoids and are also found in other fruits, such as apple, grape, and cherry. Two anthocyanins, 7-O-methylcyanidin 3-O-β-D-galactopyranoside and cyanidin 3-O-galactoside, have been identified as major anthocyanins in the skin of some mango cultivars (Berardini et al., 2005a, b; Lopez-Cobo et al., 2017). Anthocyanins are water-soluble pigments and the products of phenylalanine metabolism; this pathway has been elucidated in many plant species (Grotewold, 2006; Holton and Cornish, 1995; Saito et al., 2013; Tanaka et al., 2008). Among fruit crops, grape and apple have been most extensively investigated in research on the molecular mechanisms of anthocyanin biosynthesis (Azuma, 2018; Ban et al., 2007; Honda and Moriya, 2018; Kobayashi et al., 2004; Takos et al., 2006; Ubi et al., 2006). Boss et al. (1996a, b) reported that expression of the UDP-glucose:flavonoid 3-O-glucosyltransferase (UFGluT) gene is critical for anthocyanin biosynthesis in grape skin, and MYB transcription factors, which control UFGluT expression, are key regulators of grape skin coloration (Kobayashi et al., 2002, 2004). In apple skin, the expression of the CHS, DFR, F3H, ANS, and UFGT genes is regulated concomitant with increased anthocyanin accumulation (Honda et al., 2002), and MYB transcription factors regulating the expression of these genes have been isolated (Ban et al., 2007; Takos et al., 2006).
Light is one of the most important environmental factors that influences anthocyanin biosynthesis in fruit skin (Zoratti et al., 2014), and the molecular mechanism of anthocyanin accumulation in response to light exposure has been studied in some fruits such as grape (Azuma et al., 2012; Matus et al., 2009), apple (Feng et al., 2013; Li et al., 2012; Takos et al., 2006; Vimolmangkang et al., 2014), nectarine (Ravaglia et al., 2013) and lychee (Wei et al., 2011). Red coloration of mango skin is also affected by light exposure (Karanjalker et al., 2018a; Sivankalyani et al., 2016; Wu et al., 2013). However, the expression profile of the anthocyanin-related genes during red coloration of mango skin induced by light stimulus has not been investigated, although molecular studies of mango anthocyanin biosynthesis have been reported recently (Bajpai et al., 2018; Hoang et al., 2015; Karanjalker et al., 2018a, b). Karanjalker et al. (2018a) reported that shading or bagging fruits suppressed the expression of the anthocyanin-related genes, as well as anthocyanin content, in ripened fruit, but changes in expression levels of the genes in response to light exposure have not been investigated. As the key gene(s) or enzyme(s) regulating anthocyanin biosynthesis in mango skin have not yet been elucidated, more information on the characteristics of anthocyanin-related genes is needed to fully understand the regulation of mango skin coloration.
UDP:flavonoid 3-O-glycosyltransferase (UFGT) is a member of a large multigene family of UDP-glycosyltransferases (UGTs) and it catalyzes the final glycosylation reaction in flavonoid biosynthesis. As glycosylation of anthocyanidin increases its stability and water solubility, the expression of the UFGT gene has been associated with anthocyanin accumulation in plant species (Boss et al., 1996a, b; Hu et al., 2011; Zhao et al., 2012). To date, only a little information has been gathered about UFGT in mango. Karanjalker et al. (2018a, b) investigated expression of UFGT in mango skin, but did not publish nucleotide sequence of the gene, so no information on the sequence characteristics of mango UFGT are available. In this study, we isolated UFGT-like genes associated with anthocyanin accumulation in mango skin, and determined their expression profiles, as well as those of other anthocyanin biosynthesis-related genes, during the red coloration of mango skin.
Twenty-year-old ‘Irwin’ mango trees planted in a plastic house at the Experimental Farm of Kindai University (Yuasa town, Wakayama, Japan) were randomly selected and used in this study. In late April, about two weeks after full bloom, the tree canopy was partially covered with a light-shielding sheet (Daio Raschel 1700SG; Daio Kasei Co., Japan), which reduced the light intensity to approximately 20–25% of that of the unshielded canopy. Then, in early June, individual fruits were covered with a two-layered paper bag (Chikuma Aotake 2-7; Kobayashi Seitai Sangyo Co., Japan). To account for the possibility of the paper bag tearing as a result of fruit enlargement, each bagged fruit was covered with a larger paper bag composed of newspaper to prevent fruit coloration during the early development stage. In mid-July (July 16 in 2016; July 21 in 2017; approximately one month before harvest), the paper bags were removed to expose the fruits to sunlight in order to induce fruit coloration, while some fruits were kept bagged until harvest. Then, three bagged and three bag-removed fruits were harvested at 0, 3, 7, and 21 days after bag removal (DABR) and used to measure hue and anthocyanin content, and also for RNA extraction. The experiments were conducted for two consecutive years (2016 and 2017). RNA samples for transcriptome analysis were extracted from the red mango peel of unbagged fruits collected on June 26 in 2015. As for the tissue-specific expression analysis, the leaves (green and red), floral axes (green and red), red shoots, fruit flesh, skin of young fruit (green), skin of ripened fruit (red) and of bag-removed fruit (red) were sampled. Each tissue sample was collected at appropriate time in the 2016 season, i.e. the floral axes in late April, leaves and young fruit in late June, bag-removed fruit in late July, and ripened fruit in mid-August.
Measurement of hue (L*, a*, and b* values) and anthocyanin content of mango peelThe hue and anthocyanin content of each fruit were measured at three regions per fruit; the pedicel base area, equator area, and fruit apex area. A peel disk (10 mm diameter) was excised and the anthocyanin was extracted in 3 mL of 50% acetic acid for 72 h at 4°C in darkness. The absorbance of the extract was measured at 530 nm spectrophotometrically (UV-mini 1240; Shimadzu Co., Japan). The hue of each fruit was measured using a color difference meter (CR-400 Chroma Meter; Konica Minolta Japan, Inc., Japan).
Isolation of UFGT-like genes from mango peelTotal RNA was extracted from red mango peel by the hot borate method (Wan and Wilkins, 1994). For transcriptome analysis, library preparation, sequencing, and de novo transcriptome assembly were outsourced to Hokkaido System Science (Sapporo, Japan). In brief, a cDNA library was constructed using a TruSeq Stranded mRNA LT sample Prep Kit (Illumina Inc., CA, USA) after quality verification of the total RNA using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., CA, USA). The library was sequenced (100 bp pair-end reads) using Illumina HiSeq 2500 (Illumina Inc.). A total of 44.554 million reads were generated, and de novo assembly was performed using Trinity (Grabherr et al., 2011). Then, using the transcriptome data (S. Kanzaki, unpublished data), UFGT-like genes were screened by local blast analysis based on similarity with anthocyanin-related UFGT genes in grape (KyUFGT1; AB047090) and Arabidopsis (At5g17050; AY128739). The deduced amino acid sequence was aligned with the other plant glycosyltransferase sequences listed in Table S1 using Clustal Omega (www.clustal.org/omega/), and a phylogenetic tree was constructed using the neighbor-joining method by MEGA7 (Kumar et al., 2016) based on the aligned sequences. Based on the phylogenetic relationship with other plant glycosyltransferases, three UFGT-like genes were selected for further analysis. To clone the three UFGT-like genes, primers were designed to amplify coding regions of the genes based on the transcriptome data and used for PCR. PCR was performed with TaKaRa Ex Taq (Takara Bio Inc., Shiga, Japan) according to the manufacturer’s manual using a cDNA synthesized by a PrimeScript 1st strand cDNA Synthesis Kit (Takara Bio Inc.) from total RNA used for transcriptome analysis. Then, the amplified products were subcloned into pGEM-T Easy Vector (Promega, WI, USA), and their sequences were determined by custom sequencing service provided by Macrogen Japan (Kyoto, Japan). The obtained sequences were registered to DDBJ. To confirm the tissue-specific expression of the isolated genes, RNAs extracted from various tissues were used for expression analysis. The procedure for the expression analysis is as described below.
Expression analysis of anthocyanin biosynthesis genesPeeled mango skin was immediately frozen in liquid nitrogen and kept at −80°C until RNA extraction. Total RNA was extracted using the hot borate method (Wan and Wilkins, 1994), and then genomic DNA was removed using a Turbo DNA Free kit (Thermo Fisher Scientific, MA, USA). Total RNA was reverse transcribed using Prime Script RT Reagent kit (Takara Bio Inc.) and the transcript levels of target genes were analyzed with SYBR Premix ExTaqII (Takara Bio Inc.) on an ABI 7300 Realtime PCR System (Applied Biosystems, CA, USA) according to the manufacturer’s manual. Primers used for the expression analysis are listed in Table S2. Primers specific for structural genes of the phenylpropanoid biosynthesis pathway were designed based on the sequences reported by Hoang et al. (2015), excluding primers for MiPAL1 and MiCHI. Primers for MiPAL1, MiCHI, and transcription factors (MiMYB1, MibHLH1, MibHLH2, and MibHLH3) were designed based on homologous sequences isolated from the transcriptome data. A mango elongation factor gene (MiEF) was used as an internal control for each gene. Real-time quantitative PCR was performed in three replicates per sample, and transcript levels were normalized against MiEF. In addition, log-transformed relative gene expression in bag-removed fruits to that in bagged fruit on the same sampling date was calculated and used to make a heat map.
Obtained transcriptome data is consisted of 91,849 unigenes ranging from 224 to 11,857 bases, with an N50 length of 1,656 bases. Based on the transcriptome data, 11 unigenes were selected as UFGT-like genes through local Blast analysis (Table S3). As shown in Figure 1, the phylogenetic tree based on the deduced amino acid sequences of flavonoid UGTs was divided into five clusters. Four unigenes (TR13011, TR15975, TR37562, and TR38081) were located outside of the five clusters, indicating that these four unigenes are not associated with flavonoid glycosylation. The other seven unigenes formed clusters with previously reported flavonoid UGTs. Generally, flavonoid UGTs form a unique cluster based on their regiospecificity for sugar acceptors (Yonekura-Sakakibara et al., 2007, 2014). Three unigenes, TR59, TR9405, and TR16974, belonged to a cluster of UGTs that catalyze glycosylation at the 3-O position of flavonoid aglycones (Fig. 1). As major anthocyanins in mango skin are glycosylated at the 3-O position, we selected these three unigenes likely to be associated with anthocyanin accumulation in mango skin. In this study, TR59, TR9405, and TR16974 were named MiUFGT1, MiUFGT3, and MiUFGT4, respectively, and used for further investigation. The coding region of each gene was amplified using the primers designed based on the transcriptome data, and sequences of the amplified products were determined. The sequences of amplified MiUFGT1 and MiUFGT4 were identical to TR59 and TR9405, respectively, while MiUFGT3 showed a slight difference to TR9405. The sequences of amplified coding regions were registered to DDBJ (Accession No.: LC47860–LC47862).
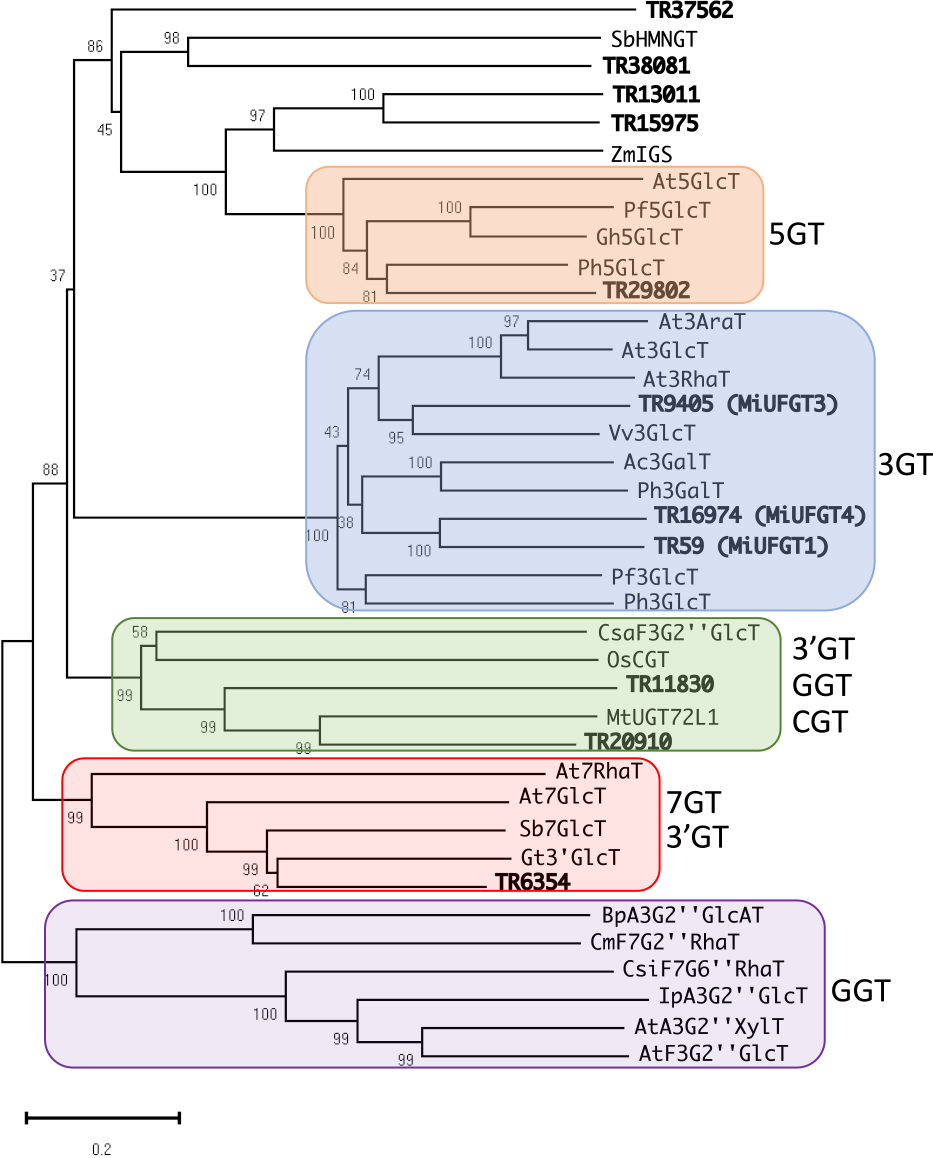
Non-rooted molecular phylogenetic tree of flavonoid glycosyltransferases and related proteins based on deduced amino acid sequences. Information on the unigenes and enzymes in this tree is shown in Tables S1 and S3. The numbers next to the branches represent the percentage of replicate trees in which the associated taxa clustered together in the bootstrap analysis (1000 replicated). Scale bar = 0.2 amino acid substitutions per site.
The deduced amino acid sequences of MiUFGT1, MiUFGT3, and MiUFGT4 showed 48, 53, and 45% identity, respectively, to the previously characterized UGT78D2 (At3GlcT in Fig. 1), a flavonoid 3-O-glucosyltransferase from Arabidopsis thaliana (Tohge et al., 2005). These three genes contain a conserved domain called the plant secondary product glycosyltransferase (PSPG) box, which was shown using three-dimensional structure analyses to interact with UDP-sugar (Gachon et al., 2005; Offen et al., 2006). Figure 2 shows alignment of the PSPG box sequences of MiUFGT1, MiUFGT3, MiUFGT4, and the UFGTs of other plant species. Interestingly, the last residue of the PSPG box in MiUFGT3 is His, which is conserved among UDP-galactose:flavonoid 3-O-galactosyltransferase (UFGalT), while in MiUFGT1 and MiUFGT4 this residue is Gln, which is conserved among UFGluTs (Kubo et al., 2004).

Alignment of the PSPG box of some flavonoid glycosyltransferases. Abbreviations for the enzymes are listed in Table S1. The black arrow indicates the amino acid residue considered to be related to the donor sugar preference of the enzyme.
Expression of the three UFGT-like genes was analyzed in several mango tissues (Fig. 3). MiUFGT3 was expressed at relatively higher levels in red tissues compared with green tissues. In a similar way, MiUFGT1 was expressed at higher levels in red leaves and floral axes compared with green leaves and floral axes, but was not detected in red peel of ripened fruit. In contrast, expression of MiUFGT4 was relatively higher in green floral axes and peel than in red floral axes and peel.

Expression of three UFGT-like genes in several tissues of ‘Irwin’ mango. Tissue samples for RNA extraction were collected at appropriate times in the 2016 season. Relative expression was analyzed via qRT-PCR and normalized to MiEF expression. GL: green leaf, RL: red leaf, GFA: green floral axis, RFP: red floral axis, RS: red shoot, F: fruit flesh, GP: green peel, RP (R): red peel of ripened fruit, RP (BR): red peel of bag-removed fruit. Values are mean ± standard error (SE; n=3).
Bagging suppressed anthocyanin accumulation in mango skin; therefore, no red coloration was observed at 0 DABR (Figs. 4, 5; Table S1). After bag removal, anthocyanin content increased rapidly and continued increasing until 21 DABR; conversely, bagged fruits did not accumulate anthocyanins (Fig. 5). Similarly, the mango skin hue changed immediately after bag removal (Table S4). The a* value increased, while the L* and b* values decreased gradually. There was little change in the skin hue of bagged fruits. In addition, the increasing a* value and anthocyanin content were higher in the pedicel base area than in the fruit apex area in bag-removed fruits. These results indicated that mango coloration is most dependent on light stimulus and intensity.
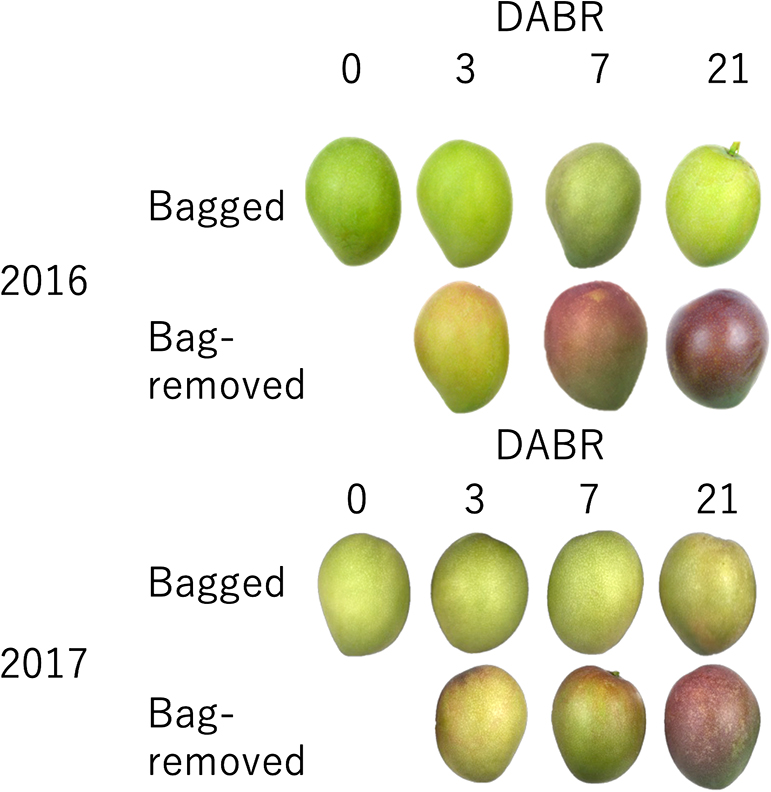
Appearance of bagged and bag-removed ‘Irwin’ mango fruits at 0, 3, 7 and 21 days after bag-removal (DABR).
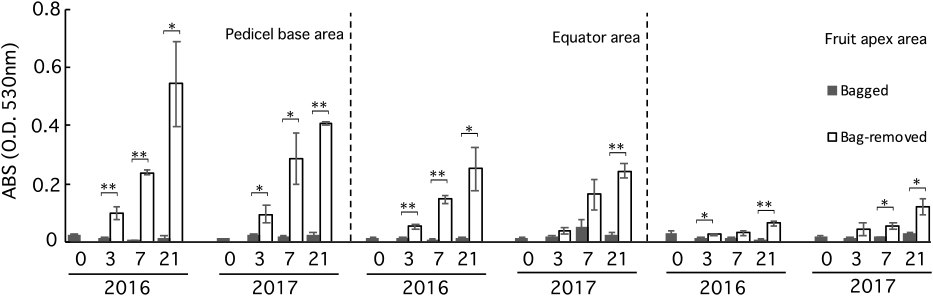
Changes in anthocyanin content in mango peel at the pedicel base area, equator area and fruit apex area of bagged (black bars) and bag-removed (white bars) fruits after bag removal. Asterisks denote significant differences according to Student’s t-test (*, P < 0.05; **, P < 0.01). Values are mean ± standard error (SE; n=3).
Genes involved in anthocyanin biosynthesis were expressed at higher levels in bag-removed fruits than in bagged fruits at 3 DABR (Figs. 6 and 7), indicating that this pathway responds rapidly to light stimulus. There were no clear differences in the expression of genes in the upper parts of the pathway, such as MiPAL1, MiPAL2, MiCHI, MiF3H, and MiF3'H, and between bagged and bag-removed fruit at 7 and 21 DABR, while MiCHS expression was significantly higher in bag-removed fruit than in bagged fruit in 2016 and 2017. The expression of MiDFR and MiANS tended to be higher in bag-removed fruit, although MiDFR had a relatively high expression level in bagged fruit in 2017. Among the three UFGT-like genes, MiUFGT4 expression was not synchronized with anthocyanin accumulation. Conversely, the expression of MiUFGT1 and MiUFGT3 was upregulated in bag-removed fruits, although this tendency was less clear in the 2017 experiments. Among the transcription factors investigated in this study, there was a clear increase in MiMYB1 expression following bag removal, parallel to anthocyanin accumulation, while there was no relationship between the expression of the three bHLH genes and mango skin coloration.

Expression profile of genes related to anthocyanin biosynthesis. Bagged (black bars) and bag-removed (white bars) fruits were collected 3, 7, and 21 DABR, while only bagged fruits were sampled on 0 DABR. Relative expression was analyzed via qRT-PCR and normalized to MiEF expression level. Asterisks denote significant differences according to Student’s t-test (*, P < 0.05; **, P < 0.01). Values are mean ± standard error (SE; n=3).
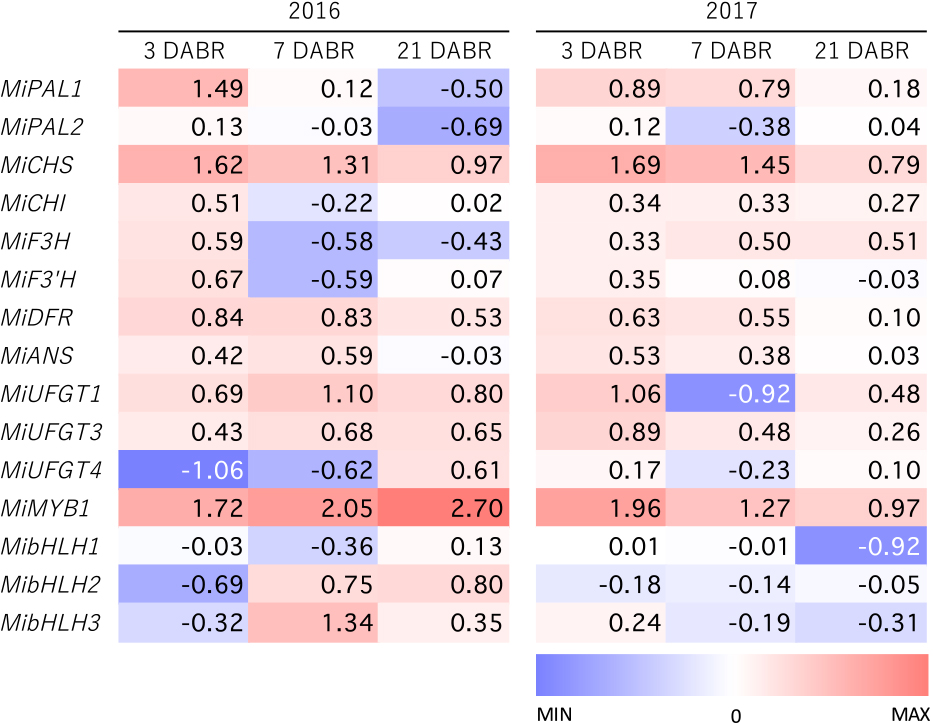
Heat map representing the expression patterns of anthocyanin-related genes. Numbers represent log-transformed relative gene expression in bag-removed fruit to that in bagged fruit on the same sampling date.
In this study, we investigated UFGT-like genes expressed in red mango skin and isolated two genes, MiUFGT1 and MiUFGT3, which are associated with anthocyanin accumulation in mango skin. To date, no sequence information on mango UFGT genes has been published, while only primer sequences for the gene were reported by Karanjalker et al. (2018a, b). Interestingly, their primers matched neither MiUFGT1 nor MiUFGT3. This indicates that MiUFGT1 and MiUFGT3 are novel genes, although this can not be confirmed until sequence information on the gene that Karanjalker et al. (2018a, b) investigated is published.
Deduced amino acid sequences of MiUFGT1 and MiUFGT3 exhibited high similarity to other plant UFGTs and contained the conserved PSPG box common to the glycosyltransferase family. The regiospecificity of UFGTs for the aglycone substrate may have arisen before speciation; therefore, UFGTs form a unique cluster in the phylogenetic tree based on the glycosylation position of sugar acceptors (Montefiori et al., 2011; Noguchi et al., 2009; Yonekura-Sakakibara et al., 2007). Consequently, the phylogenetic tree obtained in the present study indicated that MiUFGT1 and MiUFGT3 catalyze glycosylation at the 3-O position of the flavonoid aglycones (Fig. 1). MiUFGT4 also formed a cluster with MiUFGT1, MiUFGT3, and the other flavonoid 3-O-glycosyltransferases (Fig. 1); however, the expression profile of MiUFGT4 indicated that it was not involved in the coloration of mango skin (Figs. 6 and 7). The mechanism underlying the sugar donor preference of UFGTs is not well understood. Kubo et al. (2004) revealed that the presence of a histidine residue, rather than a glutamine residue, at the C-terminus end of the PSPG box appears to be conserved among galactosyltransferases, and the presence of a glutamine and a histidine residue at the C-terminus end of the PSPG box of MiUFGT1 and MiUFGT3, respectively, suggests that the products of these genes use glucose and galactose as a sugar donor, respectively. On the other hand, Offen et al. (2006) reported that the sugar donor preference was not always determined by this single amino acid substitution; therefore, the actual function and sugar donor preference of these enzymes should be elucidated by biochemical analysis. Our preliminary experiments on activity of the recombinant protein derived from MiUFGT1 and MiUFGT3 implied that the product of MiUFGT3 has galactosyltransferase activity (unpublished data). As the major anthocyanins in mango skin are galactosylated at the 3-O position of the flavonoid aglycone (Berardini et al., 2005a), it is possible that MiUFGT3 plays a significant role in anthocyanin accumulation in mango skin.
Expression analysis of MiUFGT1 and MiUFGT3 in various tissues showed that MiUFGT3 was expressed higher levels in red tissues than in green tissues (Fig. 3). MiUFGT1 was also expressed at higher levels in red leaves and floral axes compared with green leaves and floral axes. These results suggested that MiUFGT1 and MiUFGT3 may play important roles in red coloration of other tissues as well as fruit skin. Expression of MiUFGT1 was not detected in red peel of ripened fruit, but its expression was high in red peel of bag-removed fruits. A decrease in MiUFGT1 expression with fruit ripening was also observed in bag-removed fruits (Fig. 6). Therefore, expression level of MiUFGT1 may be associated with the fruit ripening stage, although further investigation will be needed to clarify this point.
In apples, bagging inhibits the coloring of fruit skin and re-exposing fruit to sunlight after bag-removal quickly induces the expression of genes involved in this process (Feng et al., 2013, 2014; Ju, 1998; Kim et al., 2003). The expression of genes involved in anthocyanin biosynthesis, such as MdMYB10, MdPAL, MdCHS, MdCHI, MdF3H, MdDFR1, MdLDOX, and MdUFGT, increased rapidly and reached maximal levels 30 hours after bag removal, and then decreased from 3 to 7 DABR, during which time anthocyanin accumulation continued (Feng et al., 2014; Kim et al., 2003). Similar results have been reported in Chinese pear (Yu et al., 2012). In the present study, gene expression was not investigated immediately after bag removal; however, the expression of anthocyanin biosynthesis-related genes at 3 DABR was higher in bag-removed fruit than in bagged fruit (Figs. 6 and 7). This suggests that gene expression increases rapidly following re-exposure of mango skin to sunlight, as observed in apple and Chinese pear. MiPAL2 and MiUFGT4 expression was not induced by bag removal, indicating that these genes are not involved in mango skin coloration. The rapid increase in expression of MiPAL1, MiCHI, MiF3H, MiF3'H, and MiDFR after bag removal indicated that these genes were up-regulated by light exposure. On the other hand, relatively high expression of these genes was observed in bagged fruit, suggesting that factors other than light may also affect the expression of these genes. In contrast, the expression of MiCHS was significantly higher in bag-removed fruit throughout the experiments in both 2016 and 2017. This indicates that MiCHS expression was strongly regulated by light stimulus. Similarly, the expression of MiANS, MiUFGT1, and MiUFGT3 tended to be controlled by light stimuli, although annual variations in the expression patterns of these genes were observed. These results suggest that MiCHS, MiANS, MiUFGT1, and MiUFGT3 may be involved in the regulation of red coloration of mango skin. As noted, the sugar donor preference of MiUFGT1 and MiUFGT3 may differ; therefore, functional analysis is needed to elucidate the roles of these genes. Regarding transcription factors, the expression of MiMYB1, which is a R2R3-MYB transcription factor with high similarity to the anthocyanin biosynthesis regulator of Arabidopsis (AtMYB90; PAP2), was observed to increase only in bag-removed fruit, indicating the light-dependent expression of MiMYB1. Therefore, it is possible that MiMYB1 is the key regulator of mango skin coloration, similar to reports in other fruit crops.
Hoang et al. (2015) and Bajpai et al. (2018) isolated anthocyanin biosynthesis pathway genes from mango and revealed that expression levels of these genes were different among cultivars with different peel colors. Additionally, Karanjalker et al. (2018a) showed that shading and bagging of mango fruits suppressed expression of anthocyanin-related genes, as well as anthocyanin accumulation, in fruit skin. However, they investigated expression of the genes only in peel of ripened fruits, so expression profiles of the genes in response to light stimulus have not been reported. In this study, we have revealed the expression patterns of the genes during red coloration induced by light exposure and showed that expression of MiCHS and MiMYB1 was strongly regulated by light stimulus. Further investigation will be needed to clarify transcription regulation of MiMYB1 in mango skin.
In conclusion, these results indicate that MiUFGT1 and MiUFGT3 are involved in the red coloration of mango skin based on their amino acid sequences and expression profiles. MiCHS and MiANS, as well as MiUFGT1 and MiUFGT3, play important roles in the anthocyanin biosynthesis of mango fruit skin and the expression of these genes is regulated by the MYB transcription factor, MiMYB1, as in other plant species. The function of mango UFGT and MYB genes should be analyzed in future studies.