2019 年 88 巻 4 号 p. 455-461
2019 年 88 巻 4 号 p. 455-461
Chromosome analysis of pineapple (Ananas comosus (L.) Merr.), one of the most important tropical fruit trees, was conducted. All experiments were carried out using root tips derived from three cross combinations as plant materials. Good preparations, with all 50 chromosomes relatively extended and well-spread without cytoplasm, were observed under enzyme conditions of “2% Cellulase Onozuka RS and 0.5% Pectolyase Y-23” without pretreatment with 2 mM 8-hydroxyquinoline. CMA-positive (+) bands were observed in telomeric positions of two chromosomes. DAPI-negative bands (−) corresponded with CMA+ bands. The numbers and positions of CMA+ bands were stable. Fluorescence in situ hybridization of rDNA was performed. The 18S-5.8S-25S rDNA sites were detected in telomeric positions of two chromosomes. The 5S rDNA sites were also detected in telomeric positions of two chromosomes. The 5S and 18S-5.8S-25S rDNA sites were located on different chromosomes. The 18S-5.8S-25S rDNA sites corresponded with the CMA+/DAPI− bands. The numbers and positions of rDNA sites were stable.
Chromosome analysis is an important procedure for genetic and biotechnological studies including breeding, genome analysis, phylogeny, and taxonomy. For this purpose, it is necessary to obtain clearly defined chromosome samples. The enzymatic maceration method (Kurata and Omura, 1978) has been effectively used to prepare chromosomes in karyological and molecular cytogenetic analyses. Some morphologically similar chromosomes have been identified by banding methods using a base-specific binding fluorochrome, guanine-cytosine (GC)-specific chromomycin A3 (CMA), and adenine-thymine (AT)-specific 4'-6-diamidino-2-phenylindole (DAPI) (Schweizer, 1976). In addition, in situ hybridization (ISH) for physical mapping of genes is a powerful tool used in advanced chromosome study. Detection of ribosomal RNA gene (rDNA) sites using fluorescence in situ hybridization (FISH) offers molecular cytogenetic information. It is essential for chromosome identification and the determining phylogenic relationships among species (Fukui et al., 1994; Murata et al., 1997).
Pineapple (Ananas comosus (L.) Merr.) is one of the most important fruit trees cultivated in tropical regions: world production was 27.4 million tons in 2017 (FAO, http://www.fao.org/faostat/en/#data/QC, June 7, 2019). The above-mentioned chromosome studies are essential to enable genetic improvements in pineapple. However, these kinds of studies have not been completed, although the chromosome number (2n = 2x = 50) and size (approx. 1 μm in metaphase) have been reported (Bhowmik, 1977; Collins and Kerns, 1931; Gitaí et al., 2005; Sharma and Ghosh, 1971). In contrast, some advanced chromosome studies were reported involving other major tropical fruit trees including banana, mango, and passionfruit (De Melo and Guerra, 2003; De Melo et al., 2001; D’Hont et al., 2000; Nishiyama et al., 2006).
Therefore, there is an urgent need to perform a study involving chromosome analysis of pineapple. Here, we conducted the following studies: 1) elucidation of conditions for an enzymatic maceration method, 2) detection of CMA− and/or DAPI-specific regions, 3) detection of 5S rDNA and 18S-5.8S-25S rDNA sites.
The plant materials used in the present study were obtained from the Nago Branch, Okinawa Prefectural Agricultural Research Center, Japan. Roots of young seedlings from seeds were obtained from three cross combinations: ‘A882’ × ‘Soft Touch’, ‘150-7-08’ × ‘Gold Barrel’, and ‘Yugafu’ × ‘Soft Touch’. Seeds were germinated in Petri dishes at 25 ± 2°C under a 14-h photoperiod with a photon flux of 57 μmol·m−2·s−1 provided by fluorescent lamps. Root tips of approximately 0.5–1 cm in length were excised. Some materials were immersed in 2 mM 8-hydroxyquinoline at 10°C for 4 h in the dark as a pretreatment, and the others did not undergo pretreatment. The root tips were fixed in methanol-acetic acid (3:1) and stored at −20°C.
Enzymatic maceration and air drying were performed as described by Fukui (1996) and Ohmido and Fukui (1996) with minor modifications. The root tips were washed in distilled water overnight at 4°C to remove the fixative. The meristematic portion of the root tips was cut on a glass slide and placed in a 1.5-mL microtube with the enzyme mixture.
For enzymatic maceration, three enzyme mixture combinations were used: 1) 2% Cellulase Onozuka RS, 0.75% Macerozyme R200 (Yakult Pharmaceutical Industry Co., Ltd., Japan), 0.15% Pectolyase Y-23 (Seishin Pharmaceutical Co., Ltd., Japan), and 1 mM EDTA, pH 4.2; 2) 4% Cellulase Onozuka RS, 1% Pectolyase Y-23, and 1 mM EDTA, pH 4.2; 3) 2% Cellulase Onozuka RS, 0.5% Pectolyase Y-23, and 1 mM EDTA, pH 4.2. The root tips were macerated at 37°C for 15–45 min.
Chromosomes were stained with 2% Giemsa solution (Merck Co., Germany) in 33.3 mM phosphate buffer (pH 6.8) for 15 min, rinsed with distilled water, air-dried, and then mounted with xylene. After confirmation of each chromosome’s position on the slide under a microscope (Nikon ECLIPSE 80i; Nicon Instech Co., Ltd., Japan), the chromosomes were destained with 70% methanol.
Fluorescent stainingFluorescent staining was performed according to the method of Hizume (1991) with minor modifications. The slides were preincubated for 30 min in McIlvaine buffer (6.6 mM citric acid and 88.2 mM Na2HPO4, pH 7.0) and treated with 0.1 g·L−1 distamycin A in the buffer for 10 min. The slides were incubated for 10 min in buffer containing 5 mM MgSO4, and then stained for 60 min with 0.1 g·L−1 CMA in buffer containing 5 mM MgSO4. The slides were also incubated for 10 min in buffer containing 5 mM MgSO4, and then stained for 30 min with 10 mg·L−1 DAPI in the buffer. The slides were incubated for 10 min in the buffer and then mounted using SlowFade (Eugene, USA). Chromosomes stained with CMA and DAPI were observed under a fluorescence microscope (ECLIPSE 80i) with a microscope digital camera (DP74, Olympus Co., Japan) using BV and UV filter cassettes, respectively.
Identification and cloning the of repeat unit of 5S rDNA and the intergenic spacerTo identify the repeat unit of pineapple 5S rDNA, a BLAST search was performed of the genome sequence of Ananas comosus (Ming et al., 2015) using the 5S rDNA sequence of Triticum aestivum (Accession number: AJ409514, DDBJ/GenBank/EMBL) (Van Campenhout et al., 2001) as a query, and setting the BLAST E-value < 1.0 × 10−20. PCR primer pairs were designed for cloning repeat units for 5S rDNA: Ac5S-F: 5'-CGGATGCGATCATACCAGCAC-3' and Ac5S-R: 5'-CCGAAAATATTGTGTCGGC-3'. PCR was performed in a 10-μL mixture containing 1× Ex Taq buffer, 2.5 mM dNTP mixture, 25 pmol of each forward and reverse primer, 5 ng genomic DNA of ‘Yonekura’, and 0.5 units of Ex Taq polymerase (Takara Bio Inc., Japan). Amplification was conducted under the following conditions: initial denaturation at 94°C for 3 min, followed by 25 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, for denaturation, annealing, and primer extension, respectively. The PCR-amplified products were separated on a 1.5% agarose gel. The 5S rDNA band was excised from the gels, and then gels were digested using Thermostable β-Agarase (Nippon Gene Co., Ltd., Japan). DNA was extracted using ethanol precipitation, cloned into the T-vector pMD20 (Takara Bio), and sequenced using an Applied Biosystems prism 3130xl genetic analyzer (Applied Biosystems, USA). The obtained sequence was registered with accession number LC458578, DDBJ/GenBank/EMBL.
rDNA sequence identification among pineapple genome sequencesSequences with high similarities to 5S rDNA and 18S-5.8S-25S rDNA were identified in the BLAST search against the pineapple genome sequence (Ming et al., 2015) using LC458578 (DDBJ/GenBank/EMBL) for 5S rDNA and X07841 (DDBJ/GenBank/EMBL) for 18S-5.8S-25S rDNA as the query and setting the BLAST E-value < 1.0 × 10−20.
Fluorescence in situ hybridization (FISH)To detect the rDNA sites, two types of rDNA probe were used in this study: 1) a 9.0-kb fragment including a full-length 18S-5.8S-25S rDNA repeat unit of wheat (Barker et al., 1988; Gerlach and Bedbrook, 1979) (accession number: X07841, DDBJ/GenBank/EMBL); 2) a 735-bp fragment of pineapple 5S rDNA (accession number: LC458578, DDBJ/GenBank/EMBL).
The 18S-5.8S-25S rDNA and 5S rDNA clones were labeled with biotin by the standard nick translation protocol (BioNick Labeling System, Invitrogen, USA). FISH was performed according to the method of Ohmido and Fukui (1996) with slight modifications (Yamamoto et al., 2010b). The biotinylated probe was hybridized to chromosomal rDNA in situ and detected with a fluorescein isothiocyanate (FITC)-avidin conjugate (Vector, California, USA) using a fluorescence microscope (ECLIPSE 80i). FITC signals were visualized using a B filter. Chromosomes were counterstained with 10 mg·L−1 DAPI in an antifadant solution (Vector Shield, Vector Laboratories, USA) and visualized using a UV filter. Signal images were analyzed using imaging software (cellSens Ver. 1.17, Olympus Co.).
To investigate the conditions of enzymatic preparation and pretreatment with 2 mM 8-hydroxyquinoline, hybrid seedlings from a combination of ‘A882’ × ‘Soft Touch’ were used as materials. Good preparations, with all 50 chromosomes relatively extended and well-spread without cytoplasm, were observed in “2% Cellulase Onozuka RS and 0.5% Pectolyase Y-23” for 35 to 45 min (Fig. 1), while some chromosomes were lost due to excessive maceration in both enzyme mixtures containing “2% Cellulase Onozuka RS, 0.75% Macerozyme R200, and 0.15% Pectolyase Y-23” and “4% Cellulase Onozuka RS and 1% Pectolyase Y-23” for 20 min. Chromosome samples with pretreatment tended to be shorter than those without pretreatment (Fig. 1). A longer chromosome sample in the prometaphase is effective for the differentiation of each chromosome. Therefore, all preparations used in subsequent chromosome analysis were prepared under the enzyme condition “2% Cellulase Onozuka RS and 0.5% Pectolyase Y-23” without pretreatment.
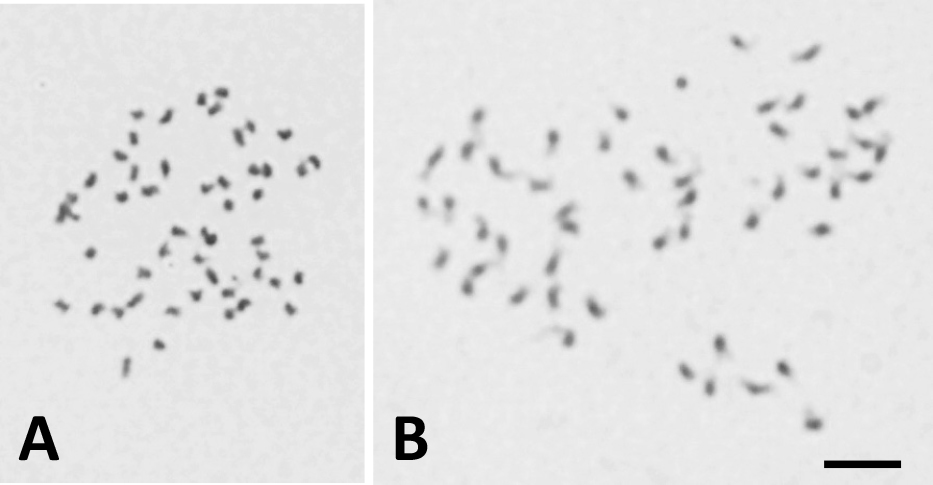
Giemsa-stained chromosomes prepared by the enzymatic maceration and air drying (EMA) method in pineapple ‘A882’ × ‘Soft Touch’ seedlings; 2% Cellulase Onozuka RS and 0.5% Pectolyase Y-23. A: pretreatment with 2 mM 8-hydroxyquinoline at 10°C for 4 h, B: without pretreatment. The bar represents 5 μm.
When CMA staining was performed, two out of 50 chromosomes exhibited CMA-positive (+) bands, chromosome regions that show bright fluorescence with CMA, and the remaining chromosomes had no CMA+ or negative (−) bands, chromosome regions that show pale fluorescence with CMA, in almost all chromosome samples derived from hybrid seedlings of the three cross combinations (Fig. 2A, C, E; Table 1). The two CMA+ bands were located in telomeric regions of the two chromosomes. These two CMA+ bands corresponded to DAPI− bands, chromosome regions that show pale fluorescence with DAPI (Fig. 2B, D, F).
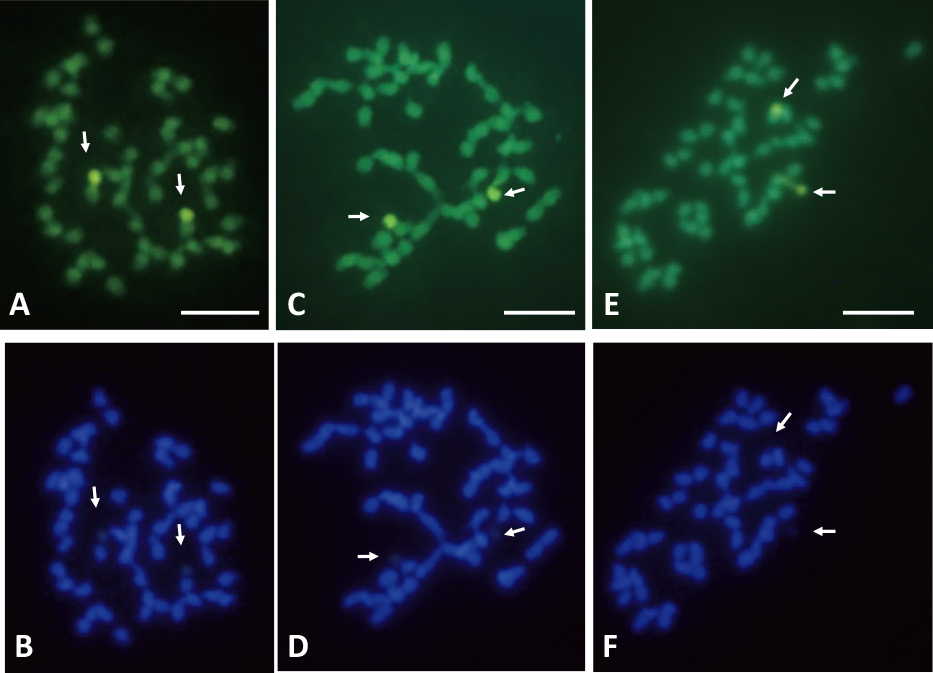
CMA- and DAPI-stained chromosomes derived from seedlings of pineapple. A and B: ‘A882’ × ‘Soft Touch’; C and D: ‘150-7-08’ × ‘Gold Barrel’; E and F: ‘Yugafu’ × ‘Soft Touch’. A, C, and E: stained with CMA; B, D, and F: stained with DAPI. Arrows indicate CMA-positive and DAPI-negative bands. Bars represent 5 μm.
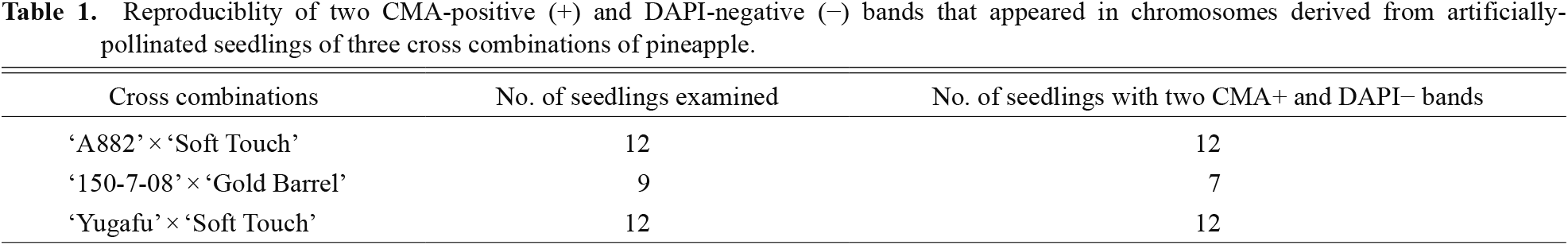
Reproduciblity of two CMA-positive (+) and DAPI-negative (−) bands that appeared in chromosomes derived from artificially-pollinated seedlings of three cross combinations of pineapple.
Identification of the 5S rDNA sequence in the pineapple genome was performed by a BLAST search using the 5S rDNA sequence of wheat as the query. This revealed that 18 of the 5S rRNA coding sequences were closely and tandemly located on Scaffold_2129 of the pineapple genome sequence (Fig. 3). Primer pairs for cloning the 5S rDNA sequence, which includes the 5S rRNA coding sequence and an intergenic spacer, were designed using the Scaffold_2129 sequence, and 5S rDNA was successfully cloned. According to a BLAST similarity search, there were 23 tandem repeats of cloned 5S rDNA in Scaffold_2129 (Fig. 3; Table 2). Other than Scaffold_2129, there were six 5S rDNA tandem repeats in LG18, five in Scaffold_1844, four in LG02, and two in LG06 and LG16. For 18S-5.8S-25S rDNA, there was one repeat unit in each of LG04, LG05, LG11, and scaffold_2500 (Table 3).

Tandem repeat of 5S rDNA and the intergenic spacer in Scaffold_2129 of the pineapple genome. A sequence region with high similarity to the cloned 5S rDNA sequence (accession number: LC458578, DDBJ/GenBank/EMBL) is shown. Arrows show 5S rDNA repeat unit. Arrowheads show the 5S rRNA coding sequence.
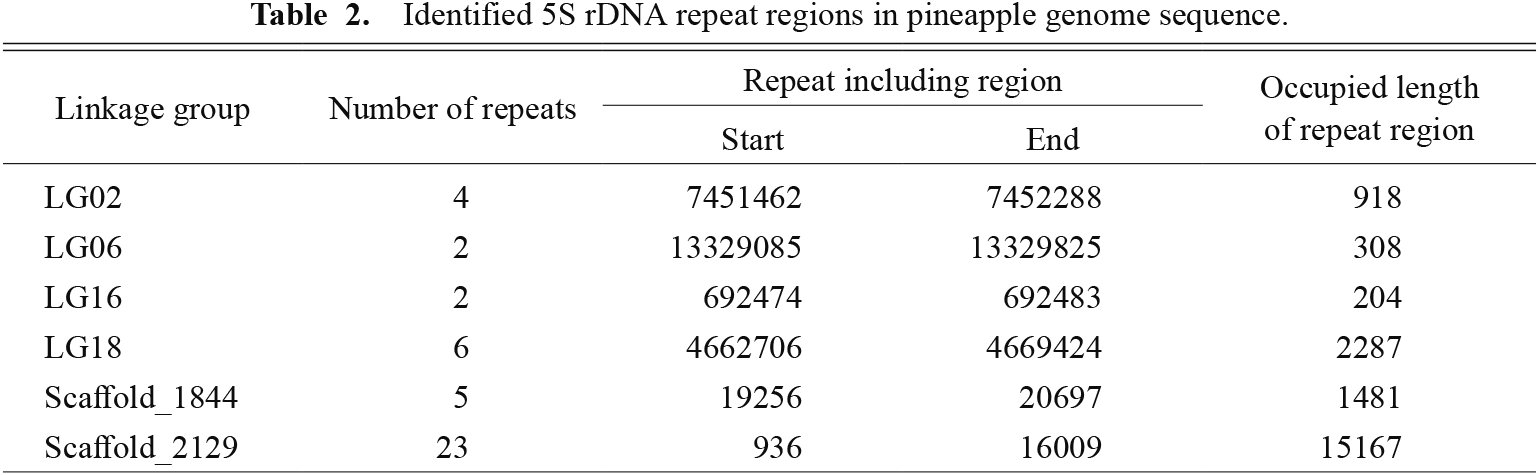
Identified 5S rDNA repeat regions in pineapple genome sequence.

Identified 18S-5.8S-25S rDNA repeat regions in pineapple genome sequence.
In situ hybridization with the 18S-5.8S-25S rDNA probe revealed signals on two chromosomes of hybrid seedlings from the combination of ‘A882’בSoft Touch’. The two signal sites were located in telomeric regions of the two chromosomes (Fig. 4B). The numbers and positions of the 18S-5.8S-25S rDNA sites were stable, and no variation among seedlings was detected (Table 4). These two chromosomes with rDNA could be easily distinguished from the other 48 chromosomes without observing rDNA sites. Two 18S-5.8S-25S rDNA sites corresponded with CMA+/DAPI− bands (Fig. 4A, B).
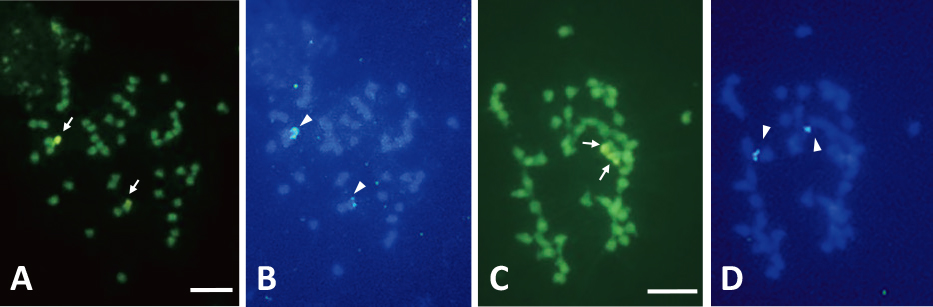
Sequential application of CMA staining and FISH of rDNA probes on somatic chromosomes of pineapple seedlings. A and B: ‘A882’ × ‘Soft Touch’; C and D: ‘Yugafu’ × ‘Soft Touch’. A and C: stained with CMA; B: FISH using 18S-5.8S-25S rDNA; D: FISH using 5S rDNA. Arrows and arrowheads indicate CMA-positive bands and rDNA sites, respectively. Bars represent 5 μm.
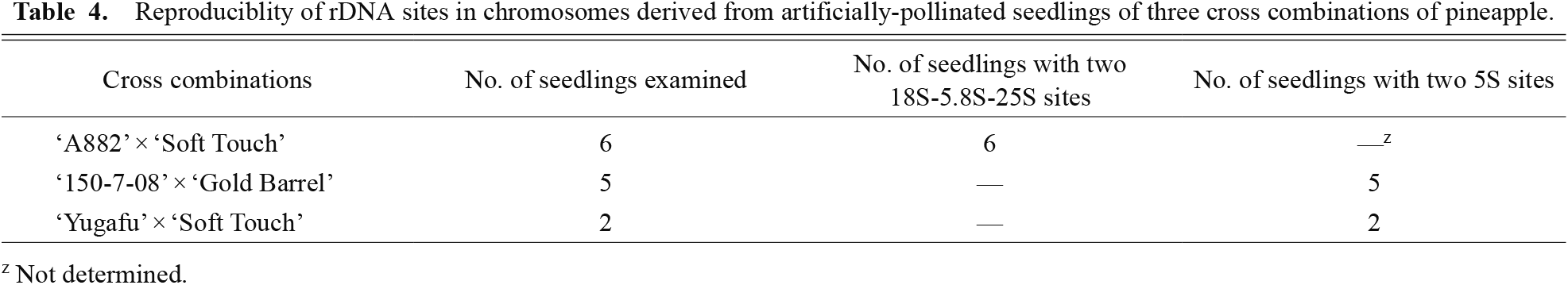
Reproduciblity of rDNA sites in chromosomes derived from artificially-pollinated seedlings of three cross combinations of pineapple.
The 5S rDNA sites were detected in telomeric positions of two chromosomes of hybrid seedlings from combinations of ‘150-7-08’בGold Barrel’ and ‘Yugafu’בSoft Touch’ (Fig. 4D). The numbers and positions of 5S rDNA sites were stable, and there was no difference among the seedlings from two cross combinations (Table 4). Two 5S rDNA and two CMA+/DAPI− bands were located on different chromosomes (Fig. 4C, D). Therefore, the 5S and 18S-5.8S-25S rDNA sites were located on different chromosomes.
In both 18S-5.8S-25S and 5S rDNA FISH, five to 15 chromosome samples of each seedling were observed. Approximately half of the samples showed two clear rDNA sites at telomeric positions of the chromosomes.
The present study demonstrated the EMA, staining of CMA and DAPI, and FISH of rDNA in pineapple chromosomes. These results provide fundamental information on pineapple chromosomes, and will contribute to the progress of cytogenetic studies.
To obtain clear results in fluorescent staining and FISH, elucidation of the conditions for obtaining good preparations, chromosomes that are relatively extended and well-spread without cytoplasm, is essential. In plants with small chromosomes, the enzymatic maceration method (Kurata and Omura, 1978) has been effectively used to prepare chromosomes in karyological and molecular cytogenetic analyses. Hence, this method has been considered promising for the small chromosomes of pineapple. For enzyme maceration of root tips of this species, “2% Cellulase Onozuka RS and 0.5% Pectolyase Y-23 at 37°C for 35 to 45 min” may be suitable. Since an adequate enzyme composition and incubation time vary depending on the materials (Fukui, 1996), elucidation of the EMA condition is considered to be crucial for the success of fluorescent staining and FISH analysis of pineapple. In addition, although pretreatment involving the collection of many mitotic chromosomes in the prometaphase or metaphase (Shishido et al., 2001), is a general technique used in plant chromosome analysis (Koba, 2006), pretreatment is not recommended for plants with small-sized chromosomes (Ohmido and Fukui, 1996). In pineapple, pretreatment is also probably unnecessary since chromosome samples with pretreatment tend to be shorter than those without pretreatment. A longer chromosome sample is desirable for differentiation of each chromosome and fluorescent staining and FISH analysis.
Fluorochrome staining analysis revealed that pineapple has two chromosomes with telomeric CMA+/DAPI− bands. The characteristics of the stains used are as follows: CMA and DAPI are GC- and AT-specific, respectively. The CMA+/DAPI− bands are considered to be GC-rich regions of chromosomes. These chromosomes with CMA+/DAPI− bands could be easily distinguished from other chromosomes without observing CMA+/DAPI− bands on CMA staining. This is the first report of CMA+/DAPI− bands in pineapple. However, a previous study (Gitaí et al., 2005) reported those regions in Aechmea bromeliifolia (Rudge) Baker, Greigia sphacelata (Ruiz & Pavon) Regel, and Ochagavia elegans R. Philippi, that belong to the same subfamily as pineapple (Bromelioideae, Bromeliaceae). Two CMA+/DAPI− bands were observed in terminal regions in G. sphacelata and O. elegans. In addition, both species possessed 2n = 2x = 50 chromosomes. Pineapple and these two species showed identical chromosome numbers and positions of CMA+/DAPI− bands. Such chromosome numbers and CMA+/DAPI− bands may be fundamental to the chromosome composition of Bromelioideae.
In the present study, we used cross-pollinated seedlings as samples. Although the genotype of each seedling was not identical to that of the parental cultivar, almost all seedlings showed the same CMA+/DAPI− bands. In addition, no cross combination difference in the CMA+/DAPI− banding pattern was observed. It can therefore be concluded that the numbers and positions of CMA+/DAPI− bands of pineapple chromosomes are very stable. This agrees with the results of peach and pear CMA/DAPI banding patterns, which showed no or few variations among seedlings and cultivars (Yamamoto et al., 1999a, 2010a). However, a few seedlings did not exhibit the two CMA+/DAPI− bands. In the present study, we could not clarify whether this was caused by genetic differences.
It is generally known that 5S rDNA contains 5S rRNA and intergenic spacers and is organized as tandem repeats in higher eukaryotic genomes. Because there were tandem repeats of 5S rRNA and intergenic spacers in the pineapple genome sequence, we identified the sequence of 5S rRNA and intergenic spacers as the 5S rDNA sequence. Although the pineapple genome sequence includes sequence information for linkage group LG01 to LG25 corresponding to the haploid chromosome number (Ming et al., 2015), BLAST analysis found most of the tandem repeats of 5S rDNA in Scaffold_2129, which is an out-group. Although small numbers of tandem repeats of 5S rDNA were found in LG02, LG06, LG16, and LG18, it is difficult to identify the locus of a 5S rDNA tandem repeat-rich region in the pineapple genome sequence. Similarly, BLAST analysis could not detect the 18S-5.8S-25S rDNA tandem repeat sequence in the pineapple genome sequence. To correspond to genomic sequence information and chromosome information detected by FISH, it is necessary to identify rDNA tandem repeat loci in the pineapple genomic sequence. However, BLAST results suggested that 5S and 18S-5.8S-25S rDNA tandem repeat region information was not fully included in the pineapple genome sequence. To identify rDNA tandem repeat loci in the pineapple genome sequence, future improvements in pineapple genome sequence information is required.
This is the first report on the number and locations of rDNA sites in pineapple. The 18S-5.8S-25S and 5S rDNA sites were located in telomeric regions of two chromosomes. The 18S-5.8S-25S and 5S rDNA sites were located on different chromosomes. The numbers and positions of two rDNA sites were stable, and no variation among seedlings was detected. The 18S-5.8S-25S rDNA sites corresponded with CMA+/DAPI− bands. This agrees with the results of peach and pear, which showed CMA+ bands that corresponded with 18S-5.8S-25S rDNA sites (Yamamoto et al., 1999b, 2012). These results indicate that 18S-5.8S-25S rDNA sites of pineapple are in a region with a high GC content (Schweizer, 1976). The 18S-5.8S-25S and 5S rDNA sites have been detected in several fruit tree species (Choi et al., 2003; Corredor et al., 2004; De Melo and Guerra, 2003; Pedrosa et al., 2000; Yamamoto et al., 2012) and their detection is useful for chromosome identification and phylogenetic studies. Therefore, the results of the present study are considered to be the first step towards applying chromosome information to these studies in pineapple.
In conclusion, we demonstrated the conditions for enzymatic maceration, CMA and/or DAPI staining, and detection of 5S rDNA and 18S-5.8S-25S rDNA sites in pineapple chromosomes. Relationships among CMA+/DAPI− bands and two types of rDNA sites were also revealed. The present results could provide fundamental information on pineapple chromosomes. Moreover, since genome studies of pineapple have advanced (Carlier et al., 2012; Ming et al., 2015; Redwan et al., 2016; Urasaki et al., 2015), studies combining genome and chromosome analyses will provide valuable information to advance studies in these and related fields. However, there are still unresolved issues. Only four out of 50 chromosomes could be identified because of the small and similar shape of chromosomes and simple CMA+/DAPI− banding pattern and rDNA sites. Since the identification of each chromosome is essential to develop breeding and genome studies, further FISH analyses using more probes (Moraes et al., 2008) and/or quantitative karyotyping (Furukawa et al., 2017) are necessary. In addition, determination of the physical locations of useful gene loci is indispensable for further progress of genome studies. The locations of useful genes on chromosomes have been elucidated in important fruit trees (Minamikawa et al., 2010; Moraes et al., 2008; Yamamoto et al., 2016). Therefore, physical mapping of useful genes, such as for leaf margin phenotypes (Urasaki et al., 2015), should be conducted in pineapple.