Abstract
Pepper yellow leaf curl disease (PepYLCD) caused by begomoviruses is one of the most devastating diseases affecting pepper (Capsicum spp.) production worldwide. In our previous study, a loss-of-function allele, pepy-1, encoding messenger RNA surveillance factor Pelota was identified as a begomovirus resistance gene from a C. annuum cultivar BaPep-5. In this study, to investigate the effectiveness of pepy-1 conferred resistance against begomovirus in the field, we conducted a three-year evaluation under natural field conditions in Indonesia. The lowest PepYLCD incidence and significantly higher fruit productivity were observed in BaPep-5 when compared to six other commonly cultivated pepper cultivars. The subsequent comparison between BaPep-5 and the susceptible BaPep-4 showed that pepy-1 slowed down the disease onset and progression, resulting in a higher fruit productivity trait in the field. Multiple comparison analyses using an F2 population obtained by crossing BaPep-5 with BaPep-4 showed that the pepy-1 homozygous individuals had significantly higher fruit productivity, twice than those of the Pepy-1 homozygous or heterozygous individuals. In conclusion, the introgression of pepy-1 is effective to reduce the economic loss of pepper fruit production under natural field infection of begomoviruses.
Introduction
Pepper (Capsicum annuum L.) is an essential condiment and vegetable crop grown in many parts of the world for innumerable purposes for food and non-food products (Tripodi and Kumar, 2019). Currently, major pepper-producing countries are China, Mexico, Turkey, Indonesia, and India, which together account for 70% of the world’s fresh and dried pepper production (FAO STAT, 2020). Indonesia produces a total of 2.7 M tonnes of fresh pepper annually to meet its domestic market’s high demand (Statistics Indonesia, 2020). Pepper production occupies more than 300,000 hectares of land in the country (Republic of Indonesia Ministry of Agriculture, 2019) indicating that it is not only an expansive horticultural commodity in the nation, but also an essential source of economic revenue.
Pepper plants are known to be vulnerable to various pathogens, including viruses, which cause enormous production losses. At present, pepper yellow leaf curl disease (PepYLCD) has been identified as one of the most destructive diseases that significantly affects global pepper production (Devendran et al., 2022). PepYLCD can be easily recognized by its distinctive typical symptoms including vein yellowing, yellow mosaic patterning, and leaf curling (Parisi et al., 2020). In severe cases, complete loss of marketable fruit has been reported (Kumar et al., 2011; Levy and Lapidot, 2008; Senanayake et al., 2012). PepYLCD is a major threat to pepper production in Indonesia and was initially identified in the western part of Java in 1999 (Hidayat et al., 1999). Since then it has rapidly spread to most parts of the country (Kesumawati et al., 2020; Sulandari et al., 2006; Tsai et al., 2009). The causal agents of PepYLCD are viruses belonging to the genus Begomovirus of the family Geminiviridae. Begomovirus comprises 445 species (ICTV, 2021), all of which have genomic structures composed of one or two circular single-stranded DNAs encapsulated in twinned icosahedral particles (Brown et al., 2015). PepYLCD is prevalent in tropical and subtropical cultivation areas including Asia, America, Africa, and the Middle East (Devendran et al., 2022), where it depends on the highly polyphagous whitefly (Bemisia tabaci Genn.) for transmission to new hosts (Gillbertson et al., 2015).
The development of begomovirus-resistant cultivars remains the most effective disease control strategy since it requires no chemical application or plant seclusion and provides an efficient and low-cost approach and a potentially stable and long-lasting solution (Lapidot and Friedmann, 2002). Breeding of begomovirus resistance is the most progressed in tomatoes (Solanum lycopersicum L.) in which several loci (Ty-1–Ty-6) against tomato yellow leaf curl virus (TYLCV) have been identified (Agrama and Scott, 2006; Anbinder et al., 2009; Gill et al., 2019; Ji et al., 2009a, b; Zamir et al., 1994), and among them, Ty-1/Ty-3, Ty-2, and ty-5 have been cloned and commercially utilized (Lapidot et al., 2015; Verlaan et al., 2013; Yamaguchi et al., 2018). In contrast, there are not yet any commercial cultivars carrying begomovirus resistance in Capsicum (Kenyon et al., 2014b). The need for adequate resistance materials is underlined by the progress in screening and the study of begomovirus resistance in Capsicum (Adluri et al., 2017; Anaya-López et al., 2003; Barchenger et al., 2019; Kenyon et al., 2014a; Mori et al., 2022; Rai et al., 2014; Retes-Manjarrez et al., 2017, 2019; Siddique et al., 2022; Singh et al., 2016; Srivastava et al., 2017; Thakur et al., 2019). Despite these germplasms being very important for breeding begomovirus resistance in commercial pepper cultivars, begomovirus resistance genes in Capsicum have only ever been identified from C. annuum BaPep-5 and PG-1 (Koeda et al., 2021, 2022), and none of these cultivars has been tested for its capacity to mitigate natural infection and minimize the damage due to yield loss under production scale.
In our previous study, we identified a recessive resistance gene pepy-1 from a C. annuum cultivar BaPep-5 for the first time in peppers and showed that this gene was effective against bipartite begomovirus pepper yellow leaf curl Indonesia virus (PepYLCIV) and pepper yellow leaf curl Aceh virus (PepYLCAV) in controlled laboratory conditions (Koeda et al., 2021). The loss-of-function allele pepy-1 has a single nucleotide polymorphism (SNP) (A to G in BaPep-5) at the splice site of the 9th intron of messenger RNA surveillance factor Pelota, which is a homolog of ty-5 in tomato. Despite this breakthrough, the effectiveness of pepy-1 in conferring resistance, as well as potential reductions in yield-loss under natural field conditions remain to be elucidated, and considering such traits will be essential for breeding new pepper cultivars. With respect to evaluating the resistance conferred by pepy-1 in reducing substantial yield loss, we conducted experiments for three consecutive years in Indonesia, an area where begomovirus is known to be severely affecting pepper cultivation.
Materials and Methods
Experimental sites, plant materials, and evaluation periods
All experiments were conducted at the experimental farm of the Agricultural Employee Training Center, Saree, Aceh Province (Northern Sumatra), Indonesia (5°26'28.0"N, 95°42'53.2"E). The field has been used to conduct PepYLCD surveys since 2012 (Koeda et al., 2016), and typical symptoms of PepYLCD, including yellowing, curling, and stunting, have been annually observed on pepper plants due to the continuously high begomovirus infection pressure. To initially compare the performance of pepy-1 conferred resistance of BaPep-5 locally called ‘Perintis’ (936 plants) with several susceptible local C. annuum cultivars, BaPep-1 ‘Top Super’ (569 plants), BaPep-2 ‘Kopay’ (597 plants), BaPep-3 ‘Odeng’ (1,762 plants), BaPep-4 ‘Kencana’ (419 plants), BaPep-6 ‘Laba’ (869 plants), and BaPep-7 ‘Kiyo’ (198 plants), yielding a total of 5,350 plants, were cultivated from December 2017 to April 2018 and designated as first-year evaluation. Consecutive evaluation of begomovirus resistance and its effect on yield was conducted by employing begomovirus resistant BaPep-5 (200 plants) and susceptible BaPep-4 (200 plants) from January to May 2019, designated as the second year. For the second year, plants of each cultivar were separated into four blocks consisting of 50 individuals each. Following the second year, to evaluate pepy-1 conferred resistance and its effect on yield parameters, 420 individuals of the F2 population derived from a cross between BaPep-5 and BaPep-4 were separated into five blocks consisting of 84 individuals each, along with a total of 25 plants of each parental line, which were separated into five blocks consisting of five plants each. The evaluation was conducted from February to July 2020 and designated as the third year.
Evaluation of begomovirus resistance
All plant materials were transplanted at the four- to the five-leaf stage and grown in rows spaced 1 m apart with a 50 cm distance between each plant and cultivated by implementing the standard local cultural practices, including irrigation, fertilization, and pesticide treatments. No PepYLCD symptoms were observed in the seedlings at transplanting. In the first year, seven pepper cultivars were evaluated every 15 days starting at 15 days post-transplantation (dpt) until 135 dpt to assess the PepYLCD incidence rate. The incidence rate percentage was determined by dividing the total number of plants showing begomovirus typical symptoms by the total number of transplanted plants for each cultivar. In the second year, BaPep-4 (susceptible) and BaPep-5 (resistant) plants were evaluated for the degree of PepYLCD symptom expression using a visual disease severity index (DSI) with scores ranging from 0 to 5 for each plant. DSI ranged from a score of 0 to 5 as follows: 0, no symptoms; 1, very mild symptoms with slight yellowing only occurring near the petioles of young leaves; 2, mild symptoms of slight yellowing of the entire area of young leaves; 3, moderate symptoms of dark yellowing of young leaves, as well as some mature leaves; 4, severe symptoms of dark yellowing of both young and most mature leaves; and 5, very severe symptoms of complete plant yellowing, stunting, and lack of fruit set. In the third year, the F2 individuals and the parental lines were evaluated using the same DSI scores.
Investigation of yield parameters
Yield parameters were recorded by collecting the mature red fruits of each tagged plant. Tagged plants for yield parameter investigation were arranged as follows: (1) in the first year: at 45 dpt, 15 representative symptomatic (early-infected) and 15 asymptomatic (late-infected) plants were tagged for each of the seven pepper cultivars, (2) in the second year: 12 representative plants from each cultivar that were symptomless at 30 dpt and became symptomatic at 45 dpt were tagged and designated as the “45 dpt-tagged plants”. Subsequently, 12 other plants from each cultivar that were symptomless at 45 dpt and became symptomatic at 60 dpt were tagged and designated as the “60 dpt-tagged plants”. Plants in both groups were divided into four blocks, each consisting of three plants from each cultivar, (3) in the third year: the F2 individuals and parent lines were tagged. Mature red fruits from each tagged plant were harvested every five days from 90 to 135 dpt to record fruit number, fruit weight (g), and fruit length (cm). The upper young leaves were collected at 135 dpt for DNA extraction.
Begomoviral DNA detection and Pepy-1 locus genotyping
DNA was extracted from collected pepper leaves using a Nucleon PhytoPure Kit (GE Healthcare, Little Chalfont, Buckinghamshire, UK) following the manufacturer’s protocol. DNA-A components of PepYLCIV, PepYLCAV, tomato yellow leaf curl Kanchanaburi virus (TYLCKaV), and ageratum yellow vein virus (AYVV) were detected using previously reported begomovirus species-specific primers (Kesumawati et al., 2019, 2020) (Supplementary Table S1). PCR was performed using EmeraldAmp MAX PCR Master Mix (Takara Bio, Kusatsu, Shiga, Japan) under the following conditions: initial denaturation at 94°C for 2 min, followed by 35 cycles at 94°C for 30 s, the annealing temperature for 30 s (Supplementary Table S1), and elongation at 72°C for 1 min. These cycles were terminated by a 3 min final extension step at 72°C. Amplified PCR products were then subjected to electrophoresis on a 1.0% (w/v) agarose gel.
Our previous study showed that an SNP (A to G) located at the splice site of the 9th intron of CaPelota in BaPep-5 (pepy-1) is responsible for resistance against PepYLCIV and PepYLCAV (Koeda et al., 2021). To genotype the Pepy-1 locus using this SNP, a high-resolution melt (HRM) analysis was conducted. Each PCR reaction contained 5 μL of Precision Melt Supermix (Bio-Rad, Hercules, CA, USA), 0.2 μM of each primer, and 2.0 μL of genomic DNA (30 ng·μL−1) in a final volume of 10 μL. PCR was performed using a CFX Connect Real-Time PCR Detection System (Bio-Rad) under the following conditions: initial denaturation at 95°C for 2 min, followed by 40 cycles of 95°C for 15 s, and 55.6°C for 20 s. HRM was run for every increment of 0.2°C between 60°C and 95°C. Primer sequences used for HRM analysis are listed in Supplementary Table S1.
Statistical analysis
To compare the total fruit yield, total fruit number, and average fruit length Student’s t-tests or Tukey–Kramer tests were performed. To compare DSI, a Mann–Whitney U-test or a Bonferroni correction test were performed. To analyze the genotypic segregation of the Pepy-1 locus in the F2 population, Pearson’s chi-square (χ2) test was performed. All statistical analyses were performed with a critical P-value of 0.05 using Excel Toukei ver. 7.0 (SSRI Limited, Tokyo, Japan).
Results
PepYLCAV is the predominant begomovirus species in the field
To confirm begomovirus infection in cultivated plants, a conventional PCR was employed to detect the predominant infecting species among PepYLCAV, PepYLCIV, TYLCKaV, and AYVV which are annually present (Koeda et al., 2016; Kesumawati et al., 2019). The result of begomovirus detection from the designated tagged plants in each year indicated (Table 1) that PepYLCD mostly contributed to the infection with PepYLCAV, which reached 100% in all samples from the first year (2018), second year (2019), followed by more than 70% of the F2 individuals in the third year (2020). Further, PepYLCIV was observed following the second-highest percentage of infection. In contrast, the infection rates of TYLCKaV and AYVV were lower compared to PepYLCAV and PepYLCIV. This finding suggested that PepYLCAV was the predominant infecting begomovirus species in the field for three years in a row.
Consistent begomovirus resistance of BaPep-5 in the multiple years
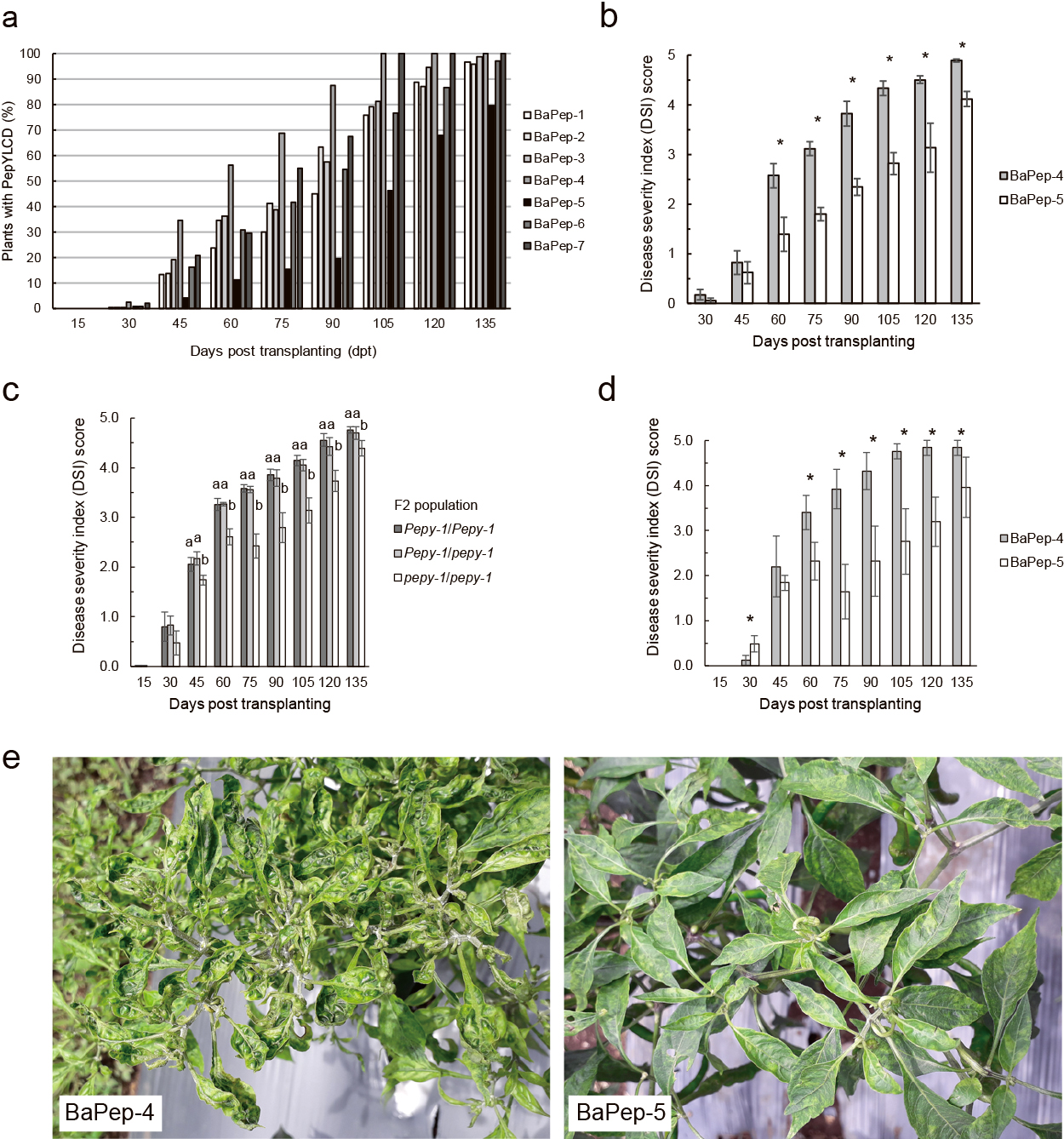
BaPep-5, together with the other seven commonly cultivated cultivars were compared for begomovirus resistance under natural field conditions in the first year. The absence of pepy-1 resistance genes in six of these cultivars (i.e., all but BaPep-5) was confirmed by HRM analysis (Supplementary Fig. S1). At 45 dpt, the highest disease incidence (35%) was found in BaPep-4, whereas disease incidence was noted to be only 4% in BaPep-5 (Fig. 1a). Disease incidence gradually increased for all pepper cultivars and reached almost 100% at 135 dpt for all cultivars except for BaPep-5, which was 80%. Based on this fact, begomovirus resistance in BaPep-5 is the most profound to be further evaluated. Based on the disease severity evaluation in the second year, BaPep-5 continuously exhibited significantly lower DSI than BaPep-4 from 60 to 135 dpt (Fig. 1b). At 75 dpt, in the reproductive phase, BaPep-4 plants showed a DSI of 3.1, while BaPep-5 plants showed a DSI of only 1.8. Based on this fact, the begomovirus resistance mechanism of BaPep-5 slows down disease onset and progression compared to susceptible cultivars.
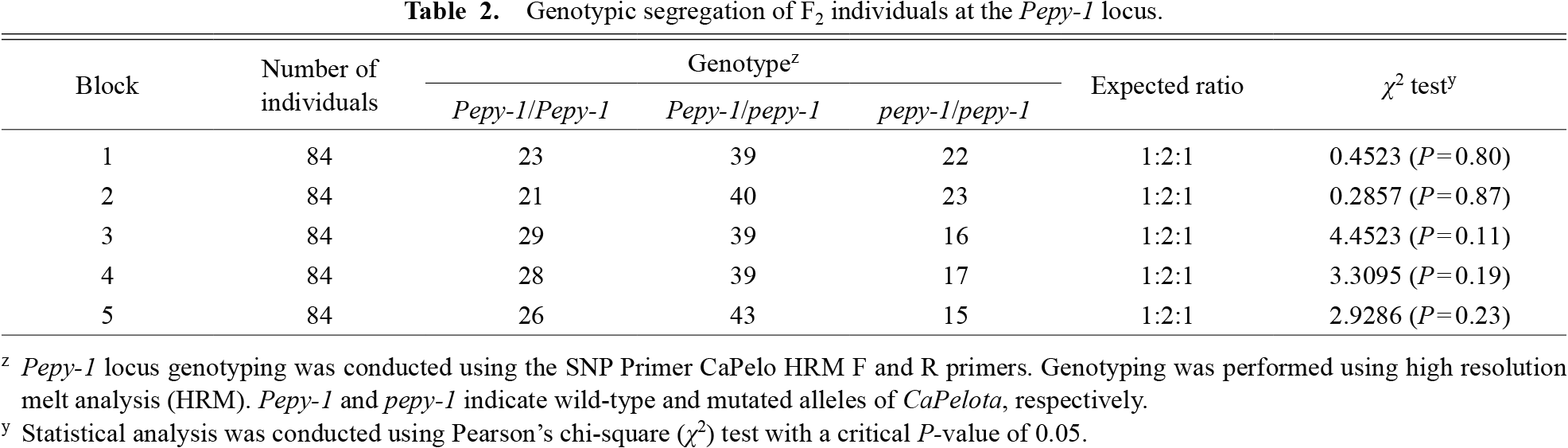
To investigate the inheritance of pepy-1, 420 F2 individuals were transplanted and separated into five blocks, each consisting of 84 plants per block. Based on the analysis, the segregation ratio of all blocks of the F2 population resulted in the expected good-fit ratio of 1:2:1 (P < 0.05) (Table 2). A total of 127 individuals were homozygous for Pepy-1, 200 individuals were heterozygous, and 93 individuals were homozygous for pepy-1. Based on this fact, the present result clarified no segregation distortion for the Pepy-1 locus, which was observed in our previous study conducted under controlled laboratory conditions (Koeda et al., 2021).
To evaluate the effectiveness of pepy-1 against begomovirus, the distribution of phenotype expression representing three genotypes (Pepy-1/Pepy-1, Pepy-1/pepy-1, and pepy-1/pepy-1) was performed in the form of DSI. As expected, the F2 individuals with a resistant genotype (pepy-1/pepy-1) showed a significantly lower average DSI than the F2 individuals with a susceptible genotype (Pepy-1/Pepy-1 and Pepy-1/pepy-1) from 45 to 135 dpt (Fig. 1c). BaPep-5 plants were resistant and BaPep-4 plants were susceptible in the same field (Fig. 1d, e). In addition, symptom recovery of the newly developed upper leaves was observed between 60 and 75 dpt in a total of 12 resistant F2 individuals (pepy-1/pepy-1); these plants showed a DSI of two at 60 dpt, which thereafter declined to zero by 75 dpt, similar to a decline in BaPep-5, where nine plants with a DSI of two at 60 dpt declined to zero at 75 dpt and one plant with a DSI of four declined to zero over the same period. Taken together, our results indicated that pepy-1 conferred resistance is effective to slow down disease onset and progression in the field.
Effect of pepy-1 conferred begomovirus resistance on fruit productivity
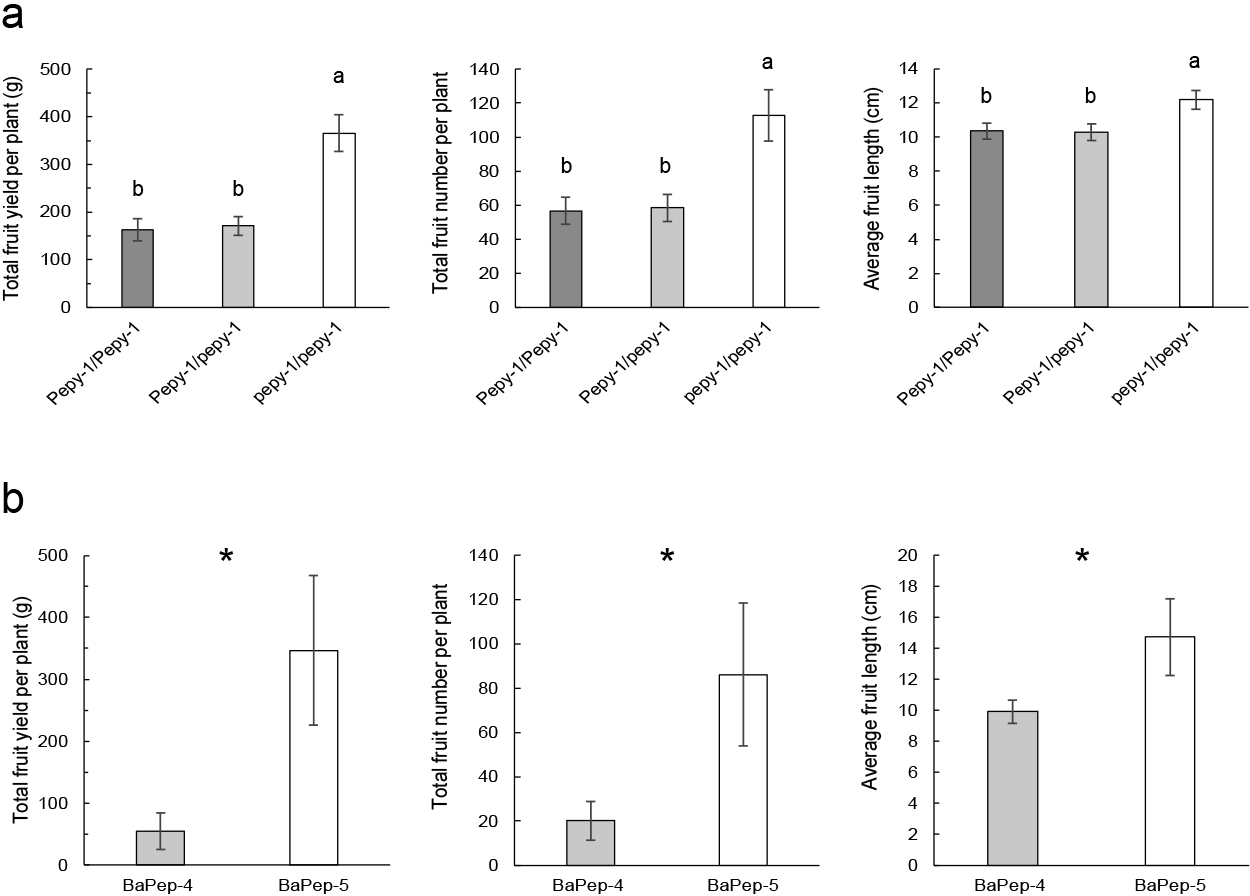
To evaluate the effect of begomovirus resistance conferred by pepy-1 in mitigating production loss, yield parameters were evaluated for three consecutive years. In the first year, multiple comparisons of three yield parameters, obtained from groups of early- and late-infected plants represented by 15 plants each, resulted in significantly higher total fruit yield per plant (g) and total fruit number harvested per plant, both in early- and late-infected BaPep-5 plants than the other six pepper cultivars (Fig. 2a). In addition, there was no reduction in the average length in early-infected plants compared to late-infected BaPep-5 plants, whereas in other cultivars plants were significantly shorter in early-infected plants, in particular BaPep-4 (Fig. 2a).

The evaluation of yield parameters was repeated in the second year by obtaining records from 45 and 60 dpt-tagged plants representing early- and late-infected plants, respectively (Fig. 2b). Total fruit yield (g) was significantly higher in both 45 and 60 dpt-tagged BaPep-5 plants compared to those of BaPep-4, with fruit yields approximately 80-fold higher for 45 dpt-tagged plants and 8-fold higher for 60 dpt-tagged plants, respectively. A significantly larger number of fruits was harvested from a single BaPep-5 plant compared to BaPep-4, with a nearly 35-fold increase at 45 dpt and 3-fold increase at 60 dpt. Not only was the average fruit length of BaPep-5 consistent between early- and late-infected plants, but it was also significantly longer than that of BaPep-4. In addition, very small fruits with no marketable value were frequently harvested from BaPep-4, indicating the extent of the difference in productivity compared to BaPep-5. Taken together, these results suggest that the stable set of large fruits in BaPep-5 under natural field conditions resulted in significantly higher fruit productivity compared to BaPep-4.
In the third year, to further investigate the effect of pepy-1 conferred begomovirus resistance on yield, we compared the yield parameters among three genotypes of the F2 population. The results showed that the F2 individuals with resistant genotype (pepy-1/pepy-1) produced a significantly higher fruit number harvested per plant and a significantly higher average fruit length than the F2 individuals with a susceptible genotype (Pepy-1/Pepy-1 and Pepy-1/pepy-1) (Fig. 3a). As a result, resistant F2 individuals accumulated a significantly higher fruit yield, approximately two times higher than the susceptible F2 individuals (Fig. 3a). These results are in accordance with the higher yield parameters of BaPep-5 plants, which were cultivated in the same field (Fig. 3b). Taken together, our data demonstrated pepy-1 conferred begomovirus resistance contributed to significantly higher fruit productivity.
Discussion
Identification of virus-resistant resources is one of the most important steps in breeding programs. In our previous study, begomovirus resistance of BaPep-5 was evaluated under controlled conditions by optimizing the artificial inoculation method for predominant begomovirus species, and resistance was shown to be controlled by pepy-1, the loss-of-function allele of messenger RNA surveillance factor Pelota (Koeda et al., 2021). Because the magnitude of disease pressure is higher in the field and environmental factors may have a strong association with resistance, natural begomovirus inoculation seemed the best approach, not only to practically evaluate the pepy-1 conferred resistance, but also to enable assessment of highly impacted yield parameters (Jiang et al., 2020; Srivastava et al., 2017). To confirm the pepy-1 conferred resistance and yield performance in the field, evaluations employing begomovirus resistant BaPep-5 and its progeny with a different set of comparisons, were repeated for three consecutive years.
The three years data set on begomovirus diagnosis results indicated that PepYLCD in the field was mostly affected by PepYLCAV, which was recently identified as a recombinant with higher virulence compared to its major parent PepYLCIV (Table 1) (Kesumawati et al., 2019; Koeda et al., 2016, 2021, 2022). Similar to PepYLCAV, earlier recombinant outbreaks with higher virulence than its putative parent have been reported from the TYLCV isolate ES421/99 in Southern Spain (Monci et al., 2002) and IS76 isolates of TYLCV-IL in Morocco (Belabess et al., 2015). Given the potential impact of recombinant virus, the continuous monitoring of PepYLCAV and its major putative parent PepYLCIV is an essential precautionary step to improve understanding of the dynamics of begomovirus populations, especially for begomovirus resistance breeding.
Multiple comparisons with several pepper cultivars were employed in the first year as they are commonly used cultivars at the local site in Indonesia. Comparing BaPep-5 to those cultivars is essential to clarify pepy-1 effectiveness to not only reduce disease incidence, but also to investigate its ability to protect the host plant from extensive production loss. The resistant BaPep-5 and the most susceptible BaPep-4 were compared in the second year to further address the pepy-1 conferred resistance to begomovirus infection, reflected by reduced disease severity established using a block design experiment. Conclusively, under the natural field inoculation of begomovirus, pepy-1 conferred resistance in BaPep-5 was effective at slowing down disease onset and progression (Fig. 1a, b).
A previous study showed that the begomovirus resistance of BaPep-5 is controlled by a single recessive gene, pepy-1, under controlled conditions, but segregation distortion was observed in the F2 population (Koeda et al., 2021). However, because normal segregations fitting to the expected ratio were observed in the F2 population in this study by optimizing natural begomovirus inoculation, it is suggested that the previous segregation distortion was most probably caused by a certain experimental bias for artificial inoculation.
PepYLCD is one of the most devastating diseases in pepper cultivation in Indonesia and other Southeast Asian countries due to its rapid transmission by whitefly and its high recombination ability, supported by the intensive and extensive cultivation of favorable host plants (Kenyon et al., 2014b). Through this field evaluation, mixed infection with multiple begomoviruses was observed (i.e., PepYLCIV, PepYLCAV, TYLCKaV, and AYVV), and more serious symptoms were observed compared to single-infected plants in controlled laboratory conditions in our previous study (Koeda et al., 2021). Such mixed infection is frequently observed in many pepper cultivating areas, and compromised resistance was reported in the C. annuum Kalyanpur Chancal cultivar under such mixed infections in the field (Singh et al., 2016). However, because BaPep-5 consistently showed significantly slower disease progression (Fig. 1), pepy-1 conferred resistance is effective, although it is not perfect, to control the disease.
Several F2 individuals with the resistant genotype showed symptom recovery in the newly developed upper leaves similar to BaPep-5 between 60 to 75 dpt in the third year (Fig. 1c, d), corresponding to our similar observation that showed restricted viral DNA accumulation under controlled conditions (Koeda et al., 2021). The symptom recovery phenomenon has been previously well-documented in the combination of bipartite begomovirus pepper golden mosaic virus (PepGMV) from Mexico and pepper plants. It was characterized as differential expressions of genes associated with plant defense machinery (Gongora-Castillo et al., 2012), following post-transcriptional gene silencing (PTGS) and transcriptional gene silencing (TGS) (Rodríguez-Negrete et al., 2009), leading to considerable symptomatic remissions of infected leaves following significantly reduced viral DNA accumulation (Carrillo-Tripp et al., 2007). A similar recovery phenomenon is also commonly observed for begomovirus-resistant genotypes in tomatoes (Bian et al., 2007; Yan et al., 2021). Our findings suggest a hypothesis of symptom reduction that could have resulted from the synergistic effect of pathways involved in TGS and PTGS, together with the putative loss of function of Pelota (pepy-1), generating enhanced resistance in BaPep-5. Notwithstanding, further study is necessary to clarify the underlying mechanism of BaPep-5 symptom recovery.
Begomovirus infection in pepper and tomato plants generally induces flower drop or abortion, ultimately resulting in a significant reduction in fruit productivity (Rojas et al., 2018). Even though yield components in pepper are qualitative traits controlled by many genes with interactions between genotype and environmental factors (Alimi et al., 2013), under such high disease pressure in the field it is most likely that virus infection severely affects yield. As higher yield-loss can be caused by infection at an earlier stage (Avilla et al., 1997; Malathi et al., 2017), the early- and late-infected plants for first- and second-year evaluations were thought to reflect the emergence day of symptoms affecting reduced fruit productivity. Based on our results, BaPep-5 was able to maintain higher fruit productivity regardless of which day the symptoms appeared (Fig. 2a, b). Further, the F2 individuals with the resistant genotype (pepy-1/pepy-1) had significantly higher fruit productivity than the F2 individuals with two susceptible genotypes (Fig. 3a). This implies that pepy-1 conferred resistance in BaPep-5 could effectively protect the host plant from significant production loss caused by begomovirus infection under production scale cultivation. Nevertheless, further study using an isogenic line is necessary to address the role of pepy-1 in fruit productivity-related traits.
Restricted viral replication caused by the putative loss-of-function mutation in Pelota was demonstrated as a novel pathway for controlling plant resistance against begomovirus infection in pepper and tomato plants (Koeda et al., 2021; Lapidot et al., 2015). Functional Pelota falls into the category of a gene facilitating a compatible interaction between a pathogen and host plants, referred to as a susceptibility (S) gene (Langner et al., 2018). In contrast to the widely exploited dominant resistance R gene, the S gene is recognized for its potential to generate non-race-specific, durable, long-lasting, and broad-spectrum disease resistance (van Schie and Takken, 2014). Many S genes are required for physiological processes in plants (Koseoglou et al., 2022); therefore, their absence resulting in loss-of-susceptibility may be accompanied by apparent pleiotropic effects as reported in Pelota mutants of tomato and rice, Mlo mutants of barley, and DND1 mutants of tomato and potato (Büschges et al., 1997; Ding et al., 2018; Lapidot et al., 2015; Qin et al., 2018; Sun et al., 2016; Zhang et al., 2018). In this study, we did not evaluate the negative pleiotropy effect of pepy-1, notably in terms of growth and yield associated traits, so this remains to be investigated in our future research.
The field evaluation of identified begomovirus resistance is essential to probe the effectiveness of the trait in actual production field conditions, notably in assessing the resistance mode against predominant isolates and its ability to reduce yield-loss, which is the most detrimental effect. To the best of our knowledge, pepy-1 and Pepy-2 are the only begomovirus resistance genes identified to date in Capsicum (Koeda et al., 2021, 2022) and therefore appropriate practical use of materials carrying these genes should be evaluated to further understanding. The present field study showed that pepy-1 conferred resistance is effective against the predominant species of PepYLCAV, PepYLCIV, TYLCKaV, and AYVV in the field, contributing to higher fruit production. Our studies showed that pepy-1 has a potential role against begomovirus under field conditions and should therefore play an important role in breeding for begomovirus resistance. To the best of our knowledge, BaPep-5 has been cultivated as a dependable local cultivar, so even if pepy-1 resulted in phenotypic pleiotropic expression, the need for its begomovirus resistance will exceed the penalty, notably in affected areas such as Indonesia. Further multilocation studies using BaPep-5 will help to clarify the effectiveness of pepy-1 conferred resistance against predominant begomovirus species from different pepper-producing areas.
Acknowledgements
We would like to thank Fitrizal and Dharma Setiawan for supporting our fieldwork in Aceh.
Literature Cited
- Adluri, P. K., G. M. Baldoldiya and P. D. Nath. 2017. Screening of Bhut Jolokia (Capsicum chinense Jacq.) germplasm of Northeast India against chili leaf curl virus. Int. J. Pure App. Biosci. 5: 1189–1196.
- Agrama, H. A. and J. W. Scott. 2006. Quantitative trait loci for Tomato yellow leaf curl virus and Tomato mottle virus resistance in tomato. J. Amer. Soc. Hort. Sci. 131: 267–272.
- Alimi, N. A., M. C. A. M. Bink, J. A. Dieleman, M. Nicolaï, M. Wubs, E. Heuvelink, J. Magan, R. E. Vooripps, J. Jansen, P. C. Rodrigues, G. W. A. M. van der Heijden, A. Vercauteren, M. Vuylsteke, Y. Song, C. Glasbey, A. Barocsi, V. Lefebvre, A. Palloix and F. A. V. Eeuwijk. 2013. Genetic and QTL analyses of yield and a set of physiological traits in pepper. Euphytica 190: 181–201.
- Anaya-López, J. L., I. Torres-Pacheco, M. González-Chavira, J. A. Garzon-Tiznado, J. L. Pons-Hernandez, R. G. Guevara-González, C. I. Muñoz-Sánchez, L. Guevara-Olvera, R. F. Rivera-Bustamante and S. Hernández-Verdugo. 2003. Resistance to geminivirus mixed infection in Mexican wild peppers. HortScience 38: 251–255.
- Anbinder, I., M. Reuveni, R. Azari, I. Paran, S. Nahon, H. Shlomo, L. Chen, M. Lapidot and I. Levin. 2009. Molecular dissection of Tomato leaf curl virus resistance in tomato line TY172 derived from Solanum peruvianum. Theor. Appl. Genet. 119: 519–530.
- Avilla, C., J. Collar, M. Duque and A. Fereres. 1997. Yield of bell pepper (Capsicum annuum) inoculated with CMV and/or PVY at different time intervals. J. Plant Dis. Prot. 104: 1–8.
- Barchenger, D. W., S. Yule, N. Jeeatid, S. W. Lin, Y. W. Wang, T. H. Lin, Y. L. Chan and L. Kenyon. 2019. A novel source of resistance to Pepper yellow leaf curl Thailand virus (PepYLCThV) (Begomovirus) in chile pepper. HortScience 54: 2146–2149.
- Belabess, Z., S. Dallot, S. El-Montaser, M. Granier, M. Majde, A. Tahiri, A. Blenzar, C. Urbino and M. Peterschmitt. 2015. Monitoring the dynamics of emergence of a non-canonical recombinant of tomato yellow leaf curl virus and displacement of its parental viruses in tomato. Virology 486: 291–306.
- Bian, X. Y., M. R. Thomas, M. S. Rasheed, M. Saeed, P. Hanson, P. J. De Barro and M. A. Rezaian. 2007. A recessive allele (tgr-1) conditioning tomato resistance to geminivirus infection is associated with impaired viral movement. Phytopathology 97: 931–937.
- Brown, J. K., F. M. Zerbini, J. Navas-Castillo, E. Moriones, R. Ramos-Sobrinho, J. C. F. Silva, E. Fiallo-Olivé, R. W. Briddon, C. Hernández-Zepeda, A. Idris, V. G. Malathi, D. P. Martin, R. Rivera-Bustamante, S. Ueda and A. Varsani. 2015. Revision of Begomovirus taxonomy based on pairwise sequence comparison. Arch. Virol. 160: 1593–1619.
- Büschges, R., K. Hollricher, R. Panstruga, G. Simons, M. Wolter, A. Frijters, R. V. Daelen, T. V. D. Lee, P. Diergaarde, J. Groenendijk, S. Töpsch, P. Vos, F. Salamini and P. Schulze-Lefert. 1997. The barley Mlo gene: A novel control element of plant pathogen resistant. Cell 88: 695–705.
- Carrillo-Tripp, J., E. Lozoya-Gloria and R. F. Rivera-Bustamante. 2007. Symptom remission and specific resistance of pepper plant after infection by Pepper golden mosaic virus. Virology. 97: 51–59.
- Devendran, R., M. Kumar, D. Ghosh, S. Yogindran, M. J. Karim and S. Chakraborty. 2022. Capsicum-infecting begomovirus as global pathogens: host-virus interplay, pathogenesis, and management. Trends Microbiol. 30: 170–184.
- Ding, W., J. Wu, J. Ye, W. Zheng, S. Wang, X. Zhu, J. Zhou, Z. Pan, B. Zhang and S. Zhu. 2018. A Pelota-like gene regulates root development and defence responses in rice. Ann. Bot. 122: 359–371.
- Food and Agriculture Organization of the United Nations (FAOSTAT). 2020. Crop and livestock product: chillies and peppers, green and dry. https://www.fao.org/faostat/en/#data/QCL/visualize.
- Gill, U., J. W. Scott, R. Shekasteband, E. Ogundiwin, C. Schuit, D. M. Francis, S. C. Sim, H. Smith and S. F. Hutton. 2019. Ty-6, a major begomovirus resistance gene on chromosome 10, is effective against Tomato yellow leaf curl virus and tomato mottle virus. Theor. Appl. Genet. 132: 1543–1554.
- Gillbertson, R. L., O. Batuman, C. G. Webster and S. Adkins. 2015. Role of the insect supervectors Bemisia tabaci and Frankliniella occidentalis in the emergence and global spread of plant viruses. Annu. Rev. Virol. 2: 67–93.
- Gongora-Castillo, E., E. Ibarra-Laclette, D. L. Trejo-Saavedra and R. F. Rivera-Bustamante. 2012. Transcriptome analysis of symptomatic and recovered leaves of geminivirus-infected pepper (Capsicum annuum). Virol. J. 9: 295. DOI: 10.1186/1743-422X-9-295.
- Hidayat, S. H., E. S. Rusli and N. Adawati. 1999. Penggunaan primer universal dalam polymerase chain reaction untuk mendeteksi virus gemini pada cabai. Prosiding Kongres Nasional XV dan Seminar Ilmiah PFI, Purwokerto, pp. 355–359.
- International Committee on Taxonomy of Viruses (ICTV). 2021. Virus taxonomy: 2021b release https://talk.ictvonline.org/taxonomy/.
- Ji, Y., J. W. Scott and D. J. Schuster. 2009a. Toward fine mapping of the Tomato yellow leaf curl virus resistance gene Ty-2 on chromosome 11 of tomato. HortScience 44: 614–618.
- Ji, Y., J. W. Scott, D. J. Schuster and D. P. Maxwell. 2009b. Molecular mapping of Ty-4, a new tomato yellow leaf curl virus resistance locus on chromosome 3 of tomato. J. Amer. Soc. Hort. Sci. 134: 281–288.
- Jiang, C., X. Wang, W. Chen, T. Liu, S. Zhong, Q. Huang, T. Ren, Z. Li, F. Tan and P. Luo. 2020. Resistance performance of wheat stripe rust resistance gene Yr41 and its effect on yield parameters in F2 populations under field conditions. Crop Prot. 134: 105168. DOI: 10.1016/j.cropro.2020.105168.
- Kenyon, L., S. Kumar, W. S. Tsai and J. d’A. Hughes. 2014a. Virus diseases of peppers (Capsicum spp.) and their control. p. 297–254. In: G. Loebenstein and N. Katis (eds.). Advances in virus research. vol. 90. Elsevier, Cambridge.
- Kenyon, L., W. S. Tsai, S. L. Shih and L. M. Lee. 2014b. Emergence and diversity of begomoviruses infecting solanaceous crop in East and Southeast Asia. Virus Res. 186: 104–113.
- Kesumawati, E., S. Okabe, K. Homma, I. Fujiwara, S. Zakaria, S. Kanzaki and S. Koeda. 2019. Pepper yellow leaf curl Aceh virus: A novel bipartite begomovirus isolated from chili pepper, tomato, and tobacco plants in Indonesia. Arch. Virol. 164: 2379–2383.
- Kesumawati, E., S. Okabe, M. Khalil, G. Alfan, P. Bahagia, N. Pohan, S. Zakaria and S. Koeda. 2020. Molecular characterization of begomoviruses associated with yellow leaf curl disease in Solanaceae and Cucurbitaceae crops from Northern Sumatra, Indonesia. Hort. J. 89: 410–416.
- Koeda, S., E. Kesumawati, Y. Tanaka, M. Hosokawa, M. Doi and A. Kitajima. 2016. Mixed infection of begomoviruses on pepper plants at Northern Sumatra, Indonesia. Trop. Agric. Dev. 60: 59–64.
- Koeda, S., N. Mori, R. Horiuchi, C. Watanabe, A. J. Nagano and H. Shiragane. 2022. PepYLCIV and PepYLCAV resistance gene Pepy-2 encodes DFDGD-Class RNA-dependent RNA polymerase in Capsicum. Theor. Appl. Genet. 135: 2437–2452.
- Koeda, S., M. Onouchi, N. Mori, N. S. Pohan, A. J. Nagano and E. Kesumawati. 2021. A recessive gene pepy-1 encoding Pelota confers resistance to begomovirus isolates of PepYLCIV and PepYLCAV in Capsicum annuum. Theor. Appl. Genet. 134: 2947–2964.
- Koseoglou, E., J. M. V. D. Wolf, R. G. F. Visser and Y. Bai. 2022. Susceptibility reversed: modified plant susceptibility genes resistance to bacteria. Trends Plant Sci. 27: 69–79.
- Kumar, S., R. Kumar, S. Kumar, A. K. Singh, M. Singh, A. B. Rai and M. Rai. 2011. Incidence of leaf curl disease on Capsicum germplasm under field condition. Indian J. Agric. Sci. 81: 187–189.
- Langner, T., S. Kamoun and K. Belhaj. 2018. CRISPR crops: plant genome editing toward disease resistance. Annu. Rev. Phytopathol. 56: 479–512.
- Lapidot, M. and M. Friedmann. 2002. Breeding for resistance to whitefly-transmitted geminivirus. Ann. Appl. Biol. 140: 109–127.
- Lapidot, M., U. Karniel, D. Gelbart, D. Fogel, D. Evenor, Y. Kutsher, Z. Makhbash, S. Nahon, H. Shlomo, L. Chen, M. Reuveni and I. Levin. 2015. A novel route controlling begomovirus resistance by the messenger RNA surveillance factor Pelota. PLoS Genet. 11: e1005538. DOI: 10.1371/journal.pgen.1005538.
- Levy, D. and M. Lapidot. 2008. Effect of plant age at inoculation on expression of genetic resistance to tomato yellow leaf curl virus. Arch. Virol. 153: 171–179.
- Malathi, V. G., P. Renukadevi, S. Chakraborty, K. K. Biswas, A. Roy, P. N. Sivalingam, V. Venkataravanappa and B. Mandal. 2017. Begomoviruses and their satellites occurring in India: distribution, diversity, and pathogenesis. p. 75–177. In: B. Mandal et al. (eds.). A century of plant virology in India. Springer, Singapore.
- Monci, F., S. Sánchez-Campos, J. Navas-Castillo and E. Moriones. 2002. A natural recombinant between the geminiviruses Tomato yellow leaf curl Sardinia virus and Tomato yellow leaf curl virus exhibits a novel pathogenic phenotype and is becoming prevalent in Spanish populations. Virology 303: 317–326.
- Mori, N., S. Hasegawa, R. Takimoto, R. Horiuchi, C. Watanabe, D. Onizaki, H. Shiragane, A. J. Nagano, E. Kesumawati and S. Koeda. 2022. Identification of QTLs conferring resistance to begomovirus isolate of PepYLCIV in Capsicum chinense. Euphytica 218: 20. DOI: 10.1007/s10681-022-02970-9.
- Parisi, M., D. Alioto and P. Tripodi. 2020. Overview of biotic stresses in pepper (Capsicum spp.): sources of genetic resistance, molecular breeding and genomics. Int. J. Mol. Sci. 21: 2587. DOI: 10.3390/ijms21072587.
- Qin, P., S. Fan, L. Deng, G. Zhong, S. Zhang, M. Li, W. Cheng, G. Wang, B. Tu, Y. Wang, X. Chen, B. Ma and S. Li. 2018. LML1, encoding a conserved eukaryotic release factor 1 protein, regulates cell death and pathogen resistance by forming a conserved complex with SPL33 in rice. Plant Cell Physiol. 59: 887–902.
- Rai, V. P., R. Kumar, S. P. Singh, S. Kumar, S. Kumar, M. Singh and M. Rai. 2014. Monogenic recessive resistance to Pepper leaf curl virus in an interspecific cross of Capsicum. Sci. Hortic. 172: 34–38.
- Republic of Indonesia Ministry of Agriculture. 2019. Data of production, harvested area, and horticulture sub-sector. https://www.pertanian.go.id/home/?show=page&act=view&id=61.
- Retes-Manjarrez, J. E., S. Hernández-Verdugo, A. Evrard and J. A. Garzón-Tiznado. 2017. Heritability of the resistance to pepper huasteco yellow vein virus in wild genotyes of Capsicum annuum. Euphytica 213: 275. DOI: 10.1007/s10681-017-2071-5.
- Retes-Manjarrez, J. E., S. Hernández-Verdugo, C. A. López-Orona, R. Medina-López, J. A. Garzón-Tiznado and J. E. Retes-Cázarez. 2019. Inheritance of resistance to Pepper huasteco yellow vein virus in Capsicum annuum L. HortScience 54: 783–786.
- Rodríguez-Negrete, E. A., J. Carrillo-Tripp and R. F. Rivera-Bustamante. 2009. RNA silencing against geminivirus: complementary action of posttranscriptional gene silencing and transcriptional gene silencing. J. Virol. 83: 1332–1340.
- Rojas, M. R., M. A. Macedo, M. R. Maliano, M. Soto-Aguilar, J. O. Souza, R. W. Briddon, L. Kenyon, R. F. Rivera Bustamante, F. M. Zerbini, S. Adkins, J. P. Legg, A. Kvarnheden, W. M. Wintermantel, M. R. Sudarshana, M. Peterschmitt, M. Lapidot, D. P. Martin, E. Moriones, A. K. Inoue-Nagata and R. L. Gilbertson. 2018. World Management of Geminiviruses. Annu. Rev. Phytopathol. 56: 637–677.
- Senanayake, D. M. J. B., A. Varma and B. Mandal. 2012. Virus–vector relationship, host range, detection and sequence comparison of Chilli leaf curl virus associated with an epidemic of leaf curl disease of chilli in Jodhpur, India. J. Phytopathol. 160: 146–155.
- Siddique, M. I., J. H. Lee, J. H. Ahn, M. K. Kusumawardhani, R. Safitri, A. Harpenas, J. K. Kwon and B. C. Kang. 2022. Genotyping-by-sequencing-based QTL mapping reveals novel loci for Pepper yellow leaf curl virus (PepYLCV) resistance in Capsicum annuum. PLoS One 17: e0264026. DOI: 10.1371/journal.pone.0264026.
- Singh, A. K., N. Kushwaha and S. Chakraborty. 2016. Synergistic interaction among begomoviruses leads to suppression of host defense-related gene expression and breakdown of resistance in chilli. Appl. Microbiol. Biotechnol. 100: 4035–4049.
- Srivastava, A., M. Mangal, R. K. Saritha and P. Kalia. 2017. Screening of chilli pepper (Capscium spp.) lines for resistance to the begomovirus causing chili leaf curl disease in India. Crop Prot. 100: 177–185.
- Statistics Indonesia. 2020. BPS—Statistics Indonesia. Production of Vegetables 2020. https://www.bps.go.id/indicator/55/61/1/produksi-tanaman sayuran.html https://www.bps.go.id/indicator/55/61/1/production-of-vegetables.html.
- Sulandari, S., R. Suseno, S. H. Hidayat, J. Harjosudarmo and S. Sosromarsono. 2006. Detection and host range study of virus associated with pepper yellow leaf curl disease. Hayati. 13: 1–6 (In Indonesian).
- Sun, K., A. M. A. Wolters, A. E. H. M. Loonen, R. P. Huibers, R. V. D. Vlugt, A. Goverse, E. Jacobsen, R. G. F. Visser and Y. Bai. 2016. Down-regulation of Arabidopsis DND1 orthologs in potato and tomato leads to broad-spectrum resistance to late blight and powdery mildew. Transgenic Res. 25: 123–138.
- Thakur, H., S. K. Jindal, A. Sharma and M. S. Dhaliwal. 2019. A monogenic dominant resistance for leaf curl virus disease in chilli pepper (Capsicum annuum L.). Crop Prot. 116: 115–120.
- Tripodi, P. and S. Kumar. 2019. The capsicum crop: an Introduction. In: N. Ramchiary and C. Kole (eds.). The capsicum genome. Compendium of plant genomes. Springer, Cham.
- Tsai, W. S., S. L. Shih, S. K. Green, L. M. Lee, G. C. Luther, M. Ratulangi, D. T. Sembel and F.-J. Jan. 2009. Identification of a new begomovirus associated with yellow leaf curl disease of tomato and pepper in Sulawesi, Indonesia. Plant Dis. 93: 321. DOI: 10.1094/PDIS-93-3-0321C.
- van Schie, C. C. and F. L. Takken. 2014. Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol. 52: 551–581.
- Verlaan, M. G., S. F. Hutton, R. M. Ibrahem, R. Komerlink, R. G. F. Visser, J. W. Scott, J. D. Edwards and Y. Bai. 2013. The tomato yellow leaf curl virus resistance genes Ty-1 and Ty-3 are allelic and code for DFDGD-Class RNA dependent RNA polymerases. PLoS Genet. 9: 1–11.
- Yamaguchi, H., J. Ohnishi, A. Saito, A. Ohyama, T. Nunome, K. Miyatake and H. Fukuoka. 2018. An NB LRR gene, TYNBS1, is responsible for resistance mediated by the Ty-2 begomovirus resistance locus of tomato. Theor. Appl. Genet. 131: 1345–1362.
- Yan, Z., A. M. A. Wolters, J. Navas-Castillo and Y. Bai. 2021. The global dimension of tomato yellow leaf curl disease current status and breeding perspectives. Microorganisms 9: 470. DOI: 10.3390/microorganisms9040740.
- Zamir, D., I. Ekstein-Michelson, Y. Zakay, N. Navot, M. Zeidan, M. Sarfatti, Y. Eshed, E. Harel, T. Pleban, H. van-Oss, N. Kedar, H. D. Rabinowitch and H. Czosnek. 1994. Mapping and introgression of a tomato yellow leaf curl virus tolerance gene, Ty-1. Theor. Appl. Genet. 88: 141–146.
- Zhang, X. B., B. H. Feng, H. M. Wang, X. Xu, Y. F. Shi, Y. He, Z. Chen, A. P. Sathe, L. Shi and J. L. Wu. 2018. A substitution mutation in OsPELOTA confers bacterial blight resistance by activating the salicylic acid pathway. J. Integr. Plant Biol. 60: 160–172.