2015 年 14 巻 3 号 p. 77-79
2015 年 14 巻 3 号 p. 77-79
We have launched a project called "Maizo"-chemistry, which is aimed toward molecular- and reaction discovery based on big data of quantum mechanical global reaction route mappings. The global reaction data includes equilibrium structures (EQs), dissociation channels (DCs), and transition structures (TSs), which are automatically calculated by a global search on a potential energy surface using the GRRM (global reaction route mapping) method. Applications to molecular- and synthesis design are an important part of the project. Machine learning and visualization techniques as well as chemoinformatics methods are essential to acquire useful information from the large reaction data space. We describe here a software system, RMapViewer, which we have developed to visualize and analyze the GRRM outputs.
The world's authority for chemical information, the CAS (Chemical Abstract Service) division of the American Chemical Society announced that more than 100 million organic and inorganic substances have been registered to their database and the pace of the registration has accelerated, and approximately 75 million were added over the past 10 years [1]. Automatic global search calculations on quantum mechanical (QM) potential energy surfaces by using the GRRM (global reaction route mapping) methods [2,3,4], however, have demonstrated that far more kinds of compounds can theoretically exist. Those unknown compounds can potentially be innovative chemical substances. We therefore have launched a project for the discovery of new molecules and reactions from the large theoretical data obtained from the QM global reaction route search. We named the project "Maizo"-chemistry. The term "Maizo [mʌizɔ:]" is Japanese, which is a prefix for buried precious items [5]. In the first stage of the project, we have developed a software system, RMapViewer, to visualize and analyze the global chemical reaction data. In the present article, we will report the development of the current version 3.0.
The enumeration of chemical structures is one of the classical topics of chemical information or chemoinformatics. A topological enumeration, such as MOLGEN [6], is the most conventional method and has been often used to generate possible chemical structures to diverse a so-called chemical space for drug discovery or structure elucidation. In the topological methods, a chemical structure is treated mathematically as a graph, which makes fast processing possible to count isomers. The topological methods are very useful but not for all cases. For example, one soon encounters a combinatorial explosion even with rather small molecular size. It will be difficult to generate chemical structures with all the relevant stereochemistry and the topological enumeration is only tractable with valence bond theory.
On the other hand, useful methods for chemical structure enumeration along with molecular orbital theory have been developed, recently. In 2004, Ohno and Maeda published the first report about the GRRM method, which makes it possible to automatically search global reaction pathways on a potential energy surface using an anharmonic downward distortion following (ADDF) search algorithm [2,3,4]. Using GRRM, one can obtain a global reaction map, which includes equilibrium structures (EQs), dissociation channels (DCs), and transition structures (TSs), together with the information about their electronic structures and energy levels along with the QM theories. Since the enumeration along with the QM theories counts structures beyond the valence bond theory as well as theoretically rational stereochemistry, it produces far more structures than the topological enumeration does. For example, in the enumeration for C6H6, the GRRM exploration has produced more than 5,000 isomers, while there are just 217 isomers in the topological basis.
The global reaction maps obtained from GRRM can provide useful data for the discovery of new molecules/reactions, synthetic design, and reaction prediction even by considering the possibilities of side reactions in the QM level. We focused on the notable characteristics of the global reaction maps and have launched a project for molecular- and reaction discovery based on the big data of the global reaction route maps. The project scheme is shown in Figure 1. Applications to molecular- and synthesis design are an important part of the project. Machine learning and visualization techniques as well as chemoinformatics methods are essential to acquire useful information from the large reaction data space. The contents and the type of computational data are different from those of experimental data. We plan to extract knowledge/models from the data and to use those knowledge/models independently or complementarily.
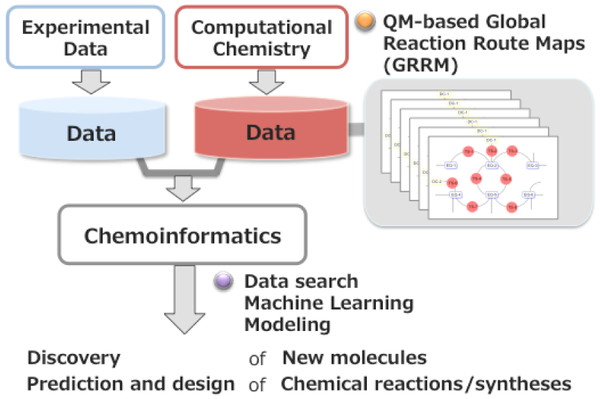
Flow of "Maizo"-chemistry project toward molecular- and reaction discovery based on the QM-based global reaction route maps (GRRM).
We assume that there are two ways of molecular/reaction discovery by using our system. One is a human-dominated way with the help of a computer, and the other is a computer-driven way. In the first stage of the project, we have developed a software system for the former way, RMapViewer, to visualize and analyze the global reaction route maps (R-maps). An example output display of RMapViewer is shown in Figure 2. In the current version of RMapViewer3.0, basic functions to analyze the R-maps have been implemented, including functions for visualization of R-maps (Figure 2–a,b), searching all possible paths between two molecules, corresponding to a reactant and a product, sorting the paths in order of TS energies and/or the number of reaction steps (Figure 2–c), and displaying a movie along a reaction coordinate. In the current version, we use the JMol tools [7] to visualize molecular models (Figure 2–d). Using the path search function (Figure 2–a,b), one can get minimum energy paths (MEPs) from reactant (s) to product (s).

Visualization and analysis of a global reaction route map using RMapViewer.
In July 2014, we released RMapViewer as freeware, which is delivered from the Sourceforge service [8]. The current version as well as the detailed usage is available with some sample input files in the RMapViewer format. We will soon release also a file converter module from the GRRM outputs to the RMapViewer inputs from the same web page.
We have been calculating the global reaction route maps for several organic molecules including C, O, N, S, H, Cl, Br, F using the GRRM system mainly at rather moderate levels of theory, such as RHF/6–31+G (d,p) and RHF/6–31+G (d). We plan to add calculations at higher theory level with larger basis sets in future. Comparing between R-maps obtained at different levels/basis sets would be also useful for the evaluation of the QM methods. Because the shape of a potential energy surface can change by a theory and/or a basis set, the number of EQ, TS, and DC structures can be changed depending on the theory/basis set used in the calculations.
In the context of the "Maizo"-chemistry project, some of the authors have found a new carbon family consisting of a prism carbon unit [9,10,11]. We also have performed conformational analyses for monosaccharides and found several MEPs between representative conformers [12].
We have given a brief introduction about a new project called "Maizo"-chemistry. Since there was no method to explore such a global range of QM potential energy surfaces before GRRM, it is expected that the large part of the global reaction route maps we have been exploring is totally new data that had not existed anywhere in the world. In the project, we plan to do mining precious findings of chemistry, which are buried in the large QM data space, by using advanced information science techniques. The global reaction data will help researchers to get an idea of molecular modeling, including in drug discovery and material design research.
The authors were supported by a Grant from the Data Centric Science Research Commons Project of the Research Organization of Information and Systems (ROIS), Japan. H.S. and T.U. were supported by a Grant-in-Aid for Challenging Exploratory Research (Grant No. 25540017) from the Japan Society for Promotion of Science.