2013 年 38 巻 2 号 p. 49-59
2013 年 38 巻 2 号 p. 49-59
Numerous weed species in many agro-ecosystems have evolved resistance to a variety of herbicides. At the present time, 388 resistant biotypes and 209 species (123 dicots and 86 monocots) in over 570,000 fields are reported.1) Thus, concerns over the sustainability of weed control have been a major driver of the development of novel herbicides and herbicide-tolerant crops. Since their first commercial introduction in 1995, the area planted by transgenic herbicide-tolerant crops has increased dramatically.2) One of the main consequences of this major change in agriculture has been the simplification of weed control; farmers can apply a single herbicide at more effective rates of the active ingredient and at multiple times during the growing season without fear of damaging the crop. However, prolonged use of a single herbicide leads to the occurrence of herbicide-resistant weeds. The most important weed-management practices are rotating or combining herbicides with different sites of action. While such practices make herbicide-tolerant crops attractive, manageability and the cost of obtaining regulatory clearance remain key issues in the production of herbicide-tolerant crops. Thus, the search is still on for better and more appropriate methods of creating herbicide-tolerant crops that depend specifically on the gene coding for an herbicide target protein.
Herbicide tolerance is provided by mutations in genes encoding the herbicide target site (target-site mutations) or genes coding for proteins that regulate the metabolism of the herbicide to a non-lethal molecule (non-target-site mutations). In the transgenic approach (Fig. 1A), herbicide tolerance is derived from random integration of a transgene with target-site mutation or non-target site mutations. In this method, herbicide-tolerant genes found in other species and organisms can be introduced into crop plants.

At present, 25% of commercialized GM breeds are herbicide-tolerant crops.3) Furthermore, transgenic soybean accounts for 47% of all the GM crop hectarage in the world, and the entire GM soybean hectarage is comprised of herbicide-tolerant varieties.2)
1.1. Commercialized herbicide-tolerant crops created by the transgenic approachMost of the increase in transgenic herbicide-tolerant crops is attributable to glyphosate-tolerant soybean, canola, cotton, maize, and sugar beet (for a review, see4)). Glyphosate competes with the substrate phosphoenolpyruvate (PEP) at the 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) enzyme-binding site in the chloroplast and inhibits the shikimate pathway. In all higher plants, EPSPS appears to be inhibited by glyphosate, making glyphosate a non-selective herbicide that is active on a very wide range of plant species. Various genetic engineering and biotechnology approaches to produce glyphosate-tolerant crops had been tried with limited success until the CP4 gene of Agrobacterium sp. was found to encode a glyphosate-tolerant form of EPSPS.5) When this CP4 gene plus a promoter was inserted into the genome of certain crops, high levels of glyphosate tolerance were expressed. Other mechanisms to deactivate glyphosate use glyphosate N-acetyltransferase (GAT). GAT acetylates glyphosate into non-phytotoxic N-acetylglyphosate.6) The GAT originally comes from Bacillus licheniformis, which has a very weak active glyphosate acetylation enzyme. Glyphosate acetylation activity was increased more than 2,000-fold by gene shuffling and site-directed mutagenesis.7,8) In addition to CP4 and GAT, a gene from Ochrobactrum anthropi encoding glyphosate oxidoreductase (GOX) was employed to deactivate glyphosate and contributed to glyphosate tolerance in canola.5)
Transgenic glufosinate-tolerant crops have also been commercialized. Glufosinate tolerance is due to glufosinate inactivation by phosphinothricin N-acetyltransferase (PAT), which catalyzes the acetylation of glufocinate.9) The bar (bialaphos resistance) gene encoding PAT were isolated from Streptomyces hygroscopicus10) and from S. viridochromogenes.11) Crop cultivars that show tolerance to both glufosinate and glyphosate are now available in corn, cotton and soybeans, and these “Dual stack” crops provide growers with a choice between two broad-spectrum herbicides and a possibility to mix the two herbicides.
Synthetic auxin herbicides control a broad spectrum of broadleaf weeds because they act rapidly on multiple receptors, competing with essential plant hormones. Different auxin receptors respond differently to different classes of auxin herbicides; thus, using the site of auxin action as a target in the design of herbicide-tolerant crops is problematic. Therefore, a transgenic approach was selected for creating auxin-herbicide-tolerant crops. A gene encoding an enzyme that deactivates an auxin herbicide was cloned from bacteria and used to generate auxin heribicide-tolerant crops. For example, a gene encoding dicamba monooxygenase (DMO, an enzyme that deactivates dicamba was cloned from the soil bacterium Stenotrophomonas maltophilla and used to generate dicamba-tolerant soybean.12,13) Other examples of auxin herbicide-tolerant crops are those expressing bacterial aryloxyalkanoate dioxygenase (AAD). AADs degrade 2,4-D and other widely used auxin herbicides effectively. While AAD-12 cleaves pyridyloxyacetate auxin herbicides such as triclopyr and fluroxypyr, AAD-1 acts on the aryloxyphenoxypropionate family of grass-active herbicides. Maize plants expressing the AAD-1 gene showed tolerance to aryloxyphenoxypropionate herbicides. Arabidopsis plants transformed with an AAD-12 gene were tolerant to 2,4-D as well as pyridyloxyacetate auxin herbicides, while tolerance to 2,4-D was maintained in transgenic soybean expressing AAD-12.14)
The enzyme 4-hydroxyphenyl pyruvate dioxygenase (HPPD) converts 4-hydroxyphenyl pyruvate to homogentisate, a key step in plastoquinone biosynthesis. Three classes of chemistry (triketones, isoxazoles, and callistemones) are HPPD-inhibiting herbicides and inhibition of HPPD causes bleaching symptoms on new growth by indirectly inhibiting carotenoid synthesis due to the requirement of plastoquinone as cofactor of phytoene desaturase (PDS).15) This is the most recently discovered herbicide mode of action, and active analogue testing continues to generate new products.16) Visible injury depends on carotenoid turnover and is, thus, slower to appear on older tissues than young leaves.17) HPPD-inhibiting herbicides control a number of important weed species and may have soil residual activity. Corn is naturally tolerant to key HPPD herbicides, but soybeans and cotton are generally sensitive. In 2011, Syngenta and Bayer Crop Science announced a co-development agreement on an HPPD herbicide-tolerance trait for soybeans18) and this trait will soon be commercialized.
Other herbicide types can also be targeted. Protoporhyrinogen oxidase (PPO) inhibiting-herbicide tolerant corn uses two point mutations at the PPO locus, PPO-1, from Arabidopsis thaliana, and rice overexpressing a naturally resistant Bacillus sublitis PPO gene exhibited tolerance to this herbicide.19) Among these transgenes, which provide herbicide tolerance, CP4, HPPD and PPO genes encode tolerant forms of herbicide target protein and GAT, GOX, BAR, DMO and AAD genes encode herbicide detoxification proteins.
1.2. Factors limiting transgenic herbicide-tolerant crop developmentHerbicide-tolerant crops created by transgenic approaches are regarded as GM crops and several factors limit the development of GM herbicide-tolerant crops, for example the high cost of obtaining regulatory clearance and international trade issues. The safety assessment required for obtaining regulatory clearance of GM crops is based on the following: 1) the presence and characteristics of newly expressed proteins and other new constituents, 2) possible alterations in the level of natural constituents beyond normal variation, and 3) characteristics of the resulting GM food and feed. Identification of these effects requires parallel molecular and phenotypic analysis of the GM plant in question and a near isogenic equivalent. In addition to the targeted analysis of specific compounds, such as macro- and micronutrients, which represent important metabolic pathways in the plant, careful analysis of the contents of known anti-nutrients and toxins was conducted. Significant differences in the transcriptome and metabolome between wild-type and transgenic plants might indicate that unintended events occur and require further investigation. GM crops are also subject to many regulatory restrictions under international trade laws. Because different countries have different review procedures, approval may not be awarded simultaneously even if all the required information is submitted at the same time, and delays can amount to years.20,21)
In contrast to the transgenic approach, mutagenesis requires chemical agents or radiation (Fig. 1B), and herbicide-tolerance is derived from spontaneous or mutagen-induced mutations in endogenous genes coding for herbicide target proteins. Herbicide-tolerant crops generated by this approach are considered as non-GM. Registration of a non-GM herbicide-tolerant crop is easier than that of a GM herbicide-tolerant crop, and non-GM herbicide-tolerant crops can obtain public acceptance more easily. Mutagenesis approaches rely on the random creation of mutations throughout the genome, and large collections of lines need to be generated to obtain the desired mutants. As the number of mutations created in a single plant is variable, an observed mutant phenotype could be the result of multi-mutations in several genes. Backcrosses with a wild-type are needed to clean up the genetic background resulting from mutagenesis.
2.1. Non-GM herbicide-tolerant crops created via mutagenesisImidazolinone (IM) herbicides inhibit acetolactate synthase (ALS), also called acetohydroxyacid synthase (AHAS), an enzyme critical for the biosynthesis of branched-chain amino acids in plants.22) ALS represents the main target for at least five structurally distinct classes of herbicides including IMs, pyrimidinylcarboxylates (PCs), sulfonylureas (SUs), triazolopyrimidine sulfonamides (TPs), and sulfonyl-aminocarbonyl-triazolinones.23) Because all commercialized IM-tolerant crops have been developed through the selection of spontaneous or mutagen-induced mutations in ALS, these crops are non-transgenic.24–28) Mutations in the ALS gene reported to confer tolerance to ALS-inhibiting herbicides are illustrated in Fig. 2.
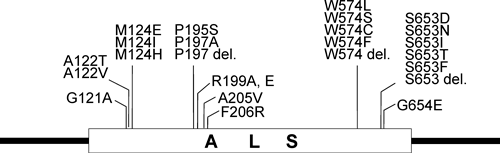
Since IM-tolerant maize was first marketed in 1992, four other IM-tolerant crops have been developed using conventional breeding methods (Table 1), now commercialized as Clearfield® crops from BASF. Because Clearfield® crops were all developed using non-GM approaches, there is no regulatory restriction on their commercialization. As a result, Clearfield® crops are more readily accessible to farmers than transgenic herbicide-tolerant crops.29)
| Plant species | Amino acid substitutionb) | Selection method |
|---|---|---|
| Maize | A122T | Pollen mutagenesis |
| W574L | Cell culture | |
| S653N | Pollen mutagenesis/Cell culture | |
| Rice | S653N | Seed mutagenesis |
| G654E | Seed mutagenesis | |
| Wheat | S653N | Seed mutagenesis |
| Oilseed rape | W574L | Microspore mutagenesis |
| S653N | Microspore mutagenesis | |
| Sunflower | A205V | Natural selection |
a) Modified from Ref. 27). b) Codon position in Arabidopsis thaliana.
Triazine-tolerant oilseed rape varieties were developed through introgression of the triazine-tolerant weed biotype cytoplasm into the triazine-susceptible oilseed rape.30) The transfer was accomplished through a combination of backcrossing and cytogenetic selection in which the triazine-tolerant weed biotype served as the donor and a susceptible canola variety served as the recurrent pollen parent.31)
Acetyl-CoA carboxylase (ACCase) is the first step of fatty acid synthesis and catalyzes the ATP (ATP)-dependent carboxylation of malonyl-CoA to form acetyl-CoA in the cytoplasm, chloroplasts, mitochondria, and peroxisomes of cells.32) ACCase-inhibiting herbicides generally inhibit the ACCase activity of monocot species and not dicots. The three chemical classes of ACCase inhibitors are cyclohexanediones (DIMs) (e.g., sethoxydim), aryloxyphenoxypropionates (FOPs) (e.g., quizalofop), and phenylpyrazolines (DENs) (e.g., pinoxaden). The first commercial ACCase-tolerant crop was a sethoxydim-tolerant corn with an altered ACCase created using tissue culture selection.33) A second ACCase trait is in the final stages of commercialization for use in sorghum. This trait was transferred with traditional breeding methods from feral sorghum (shattercane, Sorghum bicolor L. Moench), which had evolved ACCase herbicide tolerance because of agronomic practices.34)
2.2. Disadvantages of mutagenesisThe mutagenesis approach relies on the creation of random mutations throughout the whole genome; thus, it is not guaranteed that the desired mutations indeed occur in the screening population. Furthermore, if several specific mutations in the same gene are needed to acquire herbicide tolerance, it is nearly impossible to induce such mutations by this approach.
To overcome the limitations of both the transgenic and mutagenesis approaches described above, direct modification of an endogenous target gene is a major goal in crop molecular breeding. HR-dependent GT (Fig. 1C) replaces an endogenous target gene with a homologous gene with altered DNA sequence. In HR-dependent GT, a gene encoding the herbicide target protein can be replaced with an equivalent but herbicide-tolerant type of gene. HR-dependent GT has the following advantages: 1) GT can be used to replace a wild-type gene directly with its desired mutated counterpart; 2) HR-dependent GT can induce not only single-base substitutions but also multiple-base substitutions, insertions, and deletions; 3) when the difference between the endogenous and replaced gene is limited to point mutations, GT crops are genetically equivalent to non-GM crops generated by conventional mutagenesis; 4) in contrast to herbicide-tolerant crops generated by transgenic means, public acceptance of GT crops is more likely; and 5) compared to mutagenesis approaches, active induction of specific mutations can save time and costs required for obtaining preferable mutants.
3.1. Mechanism of HRHR is one of the fundamental mechanisms used to repair DNA double-strand breaks (DSBs) (Fig. 3, for a recent review, see35)). HR repairs DNA DSBs by retrieving genetic information from an undamaged homologue, such as a sister-chromatid or homologous chromosome. GT exploits HR by using extrachromosomal DNA with homology to DNA DSB regions of the plant genome as a template for retrieving a resected sequence.

Since the first report of GT of an integrated antibiotic-tolerant gene in the tobacco genome,36) various approaches aimed at HR-dependent GT have been attempted (for a review, see37–42)). Table 2 (modified from42)) summarizes HR-dependent GT attempts involving endogenous plant genes. Among successful GT reports, the strategy employed for targeting the ALS43–46) and PPO47) genes was gene-specific selection based on the fact that certain mutations in these genes could confer herbicide tolerance.
| Plant | Gene | Gene delivery system | Mutation type | Reference |
|---|---|---|---|---|
| Tobacco | ALS | Agrobacterium (protoplasts) | Point mutationb) | 43) |
| ALS | Electroporation (protoplasts) | Point mutation | 46) | |
| Arabidopsis | TGA1a-related gene 3 (TGA3) | Agrobacterium (root explants) | Knockout | 144) |
| Chalcone synthase (CHS) | Agrobacterium (suspension calli) | Knockout | 145) | |
| Agamous like 5 (AGL5) | Agrobacterium (intact plants) | Knockout | 146) | |
| Alcohol dehydrogenase (ADH) | Agrobacterium (root explants) | Knockout | 147) | |
| protoporphyrinogen oxidase (PPO) | Agrobacterium (intact plants) | Point mutation | 47) | |
| ALS | Agrobacterium (intact plants) | Point mutation | 44) | |
| Cruciferin (Cru) | Agrobacterium (intact plants) | Knockout | 104) | |
| Lotus japonicus | Glutamate-ammonia ligase (Glul), A member of Zinc finger protein (Pzf) | Agrobacterium (hypocotyl segments) | Knockout | 148) |
| Rice | Waxy | Agrobacterium (embryogenic calli) | Knockout | 149) |
| ALS | Agrobacterium (embryogenic calli) | Point mutation | 45) | |
| Alcohol dehydrogenase2 (ADH2) | Agrobacterium (embryogenic calli) | Knockout | 150) | |
| Methyltransferase1a (Met1a) | Agrobacterium (embryogenic calli) | Knockout | 151) | |
| Repressor of silencing 1 (ROS1) | Agrobacterium (embryogenic calli) | Knockin of GUS gene | 152) | |
| Domains rearranged methylase 2 (DRM2) | Agrobacterium (embryogenic calli) | Knockout | 153) | |
| α-subunit of anthranilate synthase (ASA2) | Agrobacterium (embryogenic calli) | Point mutation | 154) | |
| Maize | Inositol-1,3,4,5,6-penta-kisphosphate 2-kinase | Whisker-mediated transformation (embryogenic calli) | Knockout | 130) |
a) Modified from Ref. 40). b) Available data indicate that these are probably ectopic gene targeting events. c) Transform ZFNs with GT vector.
Endo et al.45) reported the successful creation of novel herbicide-tolerant rice plants by introducing two point mutations (W548L and S627I) into the endogenous rice ALS gene via HR-dependent GT. These point mutations correspond to W574L and S653I in the Arabidopsis ALS protein (Fig. 2). The combination of W548L and S627I mutations in rice was identified originally in suspension-cultured rice cells after a 2-year screening of bispyribac sodium (BS)-tolerant cells, through which it was found that conversion of both amino acids in ALS conferred increased tolerance to BS while each single amino acid change also conferred BS tolerance to some extent.48) However, this BS-tolerant suspension cell line lost regeneration ability due to prolonged culture and rice plants possessing these point mutations failed to regenerate.
As a template for HR, Endo et al.45) constructed a GT vector containing a partial ALS coding sequence with two point mutations; tryptophan to leucine (TGG→TTG) at amino acid 548 (W548L) and serine to isoleucine (AGT→ATT) at amino acid 627 (S627I) (Fig. 4). Because the chloroplast-targeting signal (predicted by iPSORT49)) in the 5′ region of ALS is not present in the partial ALS sequence on the T-DNA of the GT vector, random integration of T-DNA does not confer BS tolerance; the only way to produce functional and BS-tolerant ALS is for an HR event to occur between the endogenous ALS locus and the T-DNA. After co-cultivation of rice scutellum-derived calli with Agrobacterium carrying the GT vector, BS-tolerant calli were selected, and it was confirmed that both the W548L and S627I mutations had been induced in the endogenous ALS locus. GT plants homozygous for the modified ALS gene also showed hyper-tolerance to BS compared to transgenic plants over-expressing an ALS gene containing the same point mutations although the transcript level of BS-tolerant ALS GT plants is less than that of over-expressing plants. Endo et al.45) suggested that the difference might be due to the conformation of the ALS complex; Arabidopsis ALS is reported to consist of four identical subunits, similar to Saccharomyces cerevisiae ALS,50) and rice ALS has high similarity to Arabidopsis ALS. It is possible that tetramerization of ALS in plants heterozygous for the modified ALS gene combination of modified ALS with endogenous unmodified ALS or plants over-expressing an ALS gene with the same two point mutations as the modified ALS gene would be inhibited by BS. When exclusion of the BS-sensitive ALS allele is necessary to confer BS hyper-tolerance, modification of the endogenous ALS locus by GT is the most effective way to generate BS hyper-tolerant crops because an endogenous ALS locus would have been replaced with modified ALS allele by GT and subsequent crossing.

Another example of a herbicide-tolerant plant created by GT is PPO-inhibitor tolerant Arabidopsis. PPO catalyzes the last common step in heme and cholorophyll biosynthesis, i.e., oxidation for protoporphyrinogen IX to protoporphyrin.51) Inhibition of PPO by the herbicide butafenacil leads to rapid plant death; however, a combination of two specific mutations makes the PPO highly tolerant to butafenacil.52) Hanin et al.47) exploited this feature for the selection of HR-dependent GT events, with a vector containing, within the T-DNA, a 5.6-kb fragment of the Arabidopsis PPO gene modified with two mutations (S305L and Y426M) that confer butafenacil tolerance.52) Due to a deletion of the 5′ coding sequence of PPO that removes part of the chloroplast signal peptide and a nucleotide-binding motif needed for enzyme activity, the PPO gene on the GT vector is non-functional. After transformation of the GT vector to 1,400 Arabidopsis plants via influorescence infiltration,53) selection by spraying progeny seedlings with butafenacil yielded 10 tolerant individuals from a total of 4.2×106 planted seeds.
Although GT can, in principle, be used to generate any desirable property by modifying endogenous plant genes, herbicide tolerance remains the most feasible phenotype because herbicide tolerance itself can be employed as the selection marker for the GT event. Some herbicide-tolerant phenotypes are derived from a single or a few point mutations in specific genes; for genes such as ALS and PPO, GT strategies can be applied as outlined above. From this point of view, information from other herbicide-tolerant crops and weeds is very helpful for GT. Numerous weed species have evolved resistance to many herbicides in a variety of agroecosystems.1) In the search for new herbicide-resistant genes, even if specific information cannot be obtained from herbicide-tolerant crops, we can predict key amino-acid changes from weed species. Some examples of herbicide-tolerant mutations are described below.
4.1. Acetyl-CoA carboxylase (ACCase)ACCase catalyzes the first committed step of de novo fatty acid biosynthesis. Plants have both cytosolic and plastidic ACCase. In grass species, the plastidic ACCase is homometric and is the target site of three classes of herbicides. On the other hand, the plastidic ACCase in most dicots is multimeric and not sensitive to herbicides. Thus, most dicots tolerate ACCase-inhibiting herbicides well, but most grasses are susceptible to them. Originally, selectivity of aryloxy-phenoxy-propionates (APPs), cyclohexanediones (CHDs) and phyenylpyrazolinesis (PPZ) was due to different types of plastid ACCase; these herbicides interact with the carboxyl transferase domain of ACCase.54) Because of global and often intensive use of ACCase-inhibiting herbicides, many grass weeds have acquired resistance to ACCase herbicides due to amino acid substitutions in the ACCase carboxyl transferase domain (for a review, see55)); an I1781L substitution in Alopecurus myosuroides56–58) and homologous substitution in Lolium rigidum,59,60) Setaria viridis,61) Avena fatua,62) and Lolium multiflorum;63) an I2041N substitution in A. myosuroides as well as homologous substitution in L. rigidum;64) and Y2027C, G2096A, and D2078G substitutions in A. myosuroides.65) I1781L and D2078G mutations confer tolerance to both APPs and CHDs, whereas Y2027C, I2041N, and G2096A confer tolerance to APPs but not CHDs.
4.2. α-TubulinIn 1998, two research groups independently identified an α-tubulin gene mutation resulting in a T249I substitution that conferred high-level resistance to dinitroaniline in Eleusine indica.66,67) This substitution confers cross-resistance to phosphoroamidate and pyridine herbicides but increases sensitivity (negative cross-resistance) to some carbamate herbicides (for a review, see68)). The same mutation was also reported in dinitroaniline-resistant S. viridis, in which it conferred cross-resistance to benzoic acid herbicide but negative cross-resistance to carbamate herbicides.69) A second mutation, M268T, was found to provide lower-level dinitroaniline herbicide resistance in E. indica.67) Finally, an L136F mutation was also identified in dinitroaniline-resistant S. viridis.69) Fitness cost studies with plants with each of these resistance mutations are required for practical use. Target-site dinitroaniline herbicide resistance is inherited as a recessive single nuclear gene;70–74) thus, only homozygous individuals survive under herbicide treatment. If herbicide tolerance owing to mutations were inherited as a recessive trait, it would be difficult to select GT cells directly by herbicide selection because the chances of altering both alleles of the target locus by GT at once are very slim due to low GT frequency. As an alternative, it would be better to use heterozygote mutants of the target gene generated by insertional mutagenesis using transposons or Agrobacterium-mediated T-DNA insertions. If no mutants are available, we need to construct knockout mutants of the gene of interest in advance. GT can be applied to create knockout mutants as discussed in Section 7.1.
4.3. Conferring non-target site herbicide tolerance by GTALS, PPO, ACCase, and α-tubulin coding genes have been shown as candidates for conferring herbicide tolerance via GT. In addition, endogenous genes encoding an herbicide-detoxifying enzyme are candidates for conferring non-target site herbicide tolerance by GT-mediated modification. For example, glutathione S-transferases (GSTs) are families of multifunctional enzymes that catalyze the conjugation of glutathione to a variety of electrophilic and hydrophobic substrates. GSTs are involved in stress responses, and, in some crop and weed species, some herbicides are detoxified by glutathione conjugation (for reviews, see75–77)). Herbicide-metabolizing GSTs have been purified and characterized from several crops (for reviews, see75,78)). Structure analysis (crystal analysis), molecular modeling and mutagenesis studies provide an understanding of the molecular basis of GST-catalyzed herbicide binding and how single amino acid substitution(s) can improve GST catalytic efficiency and affect substrate specificity for herbicides and xenobiotics.79–81)
GOX, GAT, DMO, and ADD genes used for creating herbicide-tolerant crops also encode non-target site herbicide-tolerant proteins, but these transgenes are derived from microorganisms. When endogenous counterparts of these genes in crops are revealed and the herbicide metabolic mechanism is clarified, GT can be applied to create non-target site herbicide-tolerant crops by modifying these endogenous genes.
A major advantage of HR-dependent GT is the precise modification of the target gene. However, the risks of unintended genome modification that accompany tissue culture, namely, Agrobacterium infection and transformation with the GT vector, remain. Indeed, unexpected gene modifications accompanying GT have been reported; ectopic GT events, in which the GT product seems to be integrated elsewhere in the genome, were detected in several GT studies.43,44,47) In ectopic GT, genomic DNA is copied from the target locus to the T-DNA, and this synthesized T-DNA is integrated elsewhere in the genome.44) Another concern is random integration of the GT vector. Although Agrobacterium-mediated transformation is useful for plant transformation because this method can deliver T-DNA into plant nuclei efficiently, GT and random integration of T-DNA (GT vector) can occur simultaneously in the same cell and often results in unwanted, high-copy-number, T-DNA integration.82–85) An additional problem with Agrobacterium-mediated transformation is the propensity for DNA sequences outside the T-DNA region to integrate into the plant genome.84,86,87) For example, Kononov et al.86) detected the backbone sequence in 75% of tested transgenic tobacco (Nicotiana tabacum) plants, and the presence of the entire vector backbone very often presents potential troubles for the plant genome.88) T-DNA vector backbones usually harbor bacterial antibiotic resistance genes, which can lead to regulatory concerns. Furthermore, integration of Agrobacterium chromosomal DNA into the plant genome has been reported.89,90)
PCR analysis, Southern blot analysis, oligo array (also known as DNA chip), and whole genome sequencing are currently used for analyzing modifications to the genome. PCR-based methods require only small amounts of plant material and are easily automated for high-throughput quantitation. In Southern analysis, a blot of digested genomic plant DNA is hybridized with a transgene probe to yield an informative band pattern. With the appropriate choice of a restriction enzyme and a probe sequence, integration of a partial vector sequence might be detected. Oligo arrays allow the analysis of multiple sequence targets in a single assay. Designing an oligo array with a set of DNA probes covering the whole vector sequence, and hybridizing genomic DNA of wild-type and herbicide-tolerant crops created by GT may reveal the integration of partial sequences of the GT vector. Whole-genome sequencing is the most precise way to reveal the genome modifications in GT crops. Sequencing technologies have been developing rapidly, and information can be obtained not only about the insertion of the vector sequence, but also about the somaclonal mutations that occur during tissue culture. We anticipate that sequencing of whole genomes will become a standard technique in the characterization of transgenic and GT crops in the near future.
HR-dependent GT is a powerful tool for modifying endogenous plant genes in an expected and predictable manner. However, a still recalcitrant problem with GT is its low frequency. In fact, when we transform with a GT vector with homology to a target gene, almost all T-DNAs integrate randomly into the plant genome, and HR between the target gene and the GT vector occurs only rarely. The frequency of HR-dependent GT has been shown to be in the order of 10−3 to 10−6 relative to the random integration of the GT vector.39,42,43,91–97) Various approaches have been taken to overcome the difficulties in performing efficient GT. One such approach is the improvement of selection efficiency and another is the improvement of HR efficiency.
7.1. Improvement of selection efficiency using a positive-negative selection markerAs reported above (Section 4.2.), if herbicide tolerance owing to mutations is inherited as a recessive trait, it would be better to use heterozygous mutants of the target gene. A GT system using a positive-negative selection marker (Fig. 5), which was originally developed to enrich rare knockout mutants in mouse,98) is available for generating knock-out and knock-in mutants. In positive-negative selection vectors, positive selection markers are located within segments homologous to the target genes, and negative selection markers flank the targeting homologous sequence, acting to counter-select for random or non-targeted integration events (Fig. 5A). In higher plants, the NPTII gene for resistance to kanamycin or geneticin (G418) and the HPT gene for hygromycin resistance have been used as a positive selection marker. The codA gene encoding cytosine deaminase and the DT-A gene encoding the diphtheria toxin A fragment were used as negative selection markers because these genes confer lethal or conditional lethal phenotypes (for a review, see42)). As an aside, positive-negative selection is useful not only for generating knock-out mutants of specific genes but also for introducing point mutations that eliminate the positive selection marker after GT (Fig. 5B). There are several strategies to exclude selection marker genes, such as site-specific recombination,99,100) transposition systems,101) and HR.102,103) The combination of positive-negative selection-dependent GT and subsequent elimination of the positive selection marker gene can be applied to the induction of mutations in any gene if the induced mutation itself does not provide a selectable phenotype. If herbicide tolerance acquired by point mutations is not sufficiently strong at the tissue selection stage of GT, such a tandem approach may be successful to insert point mutations at the desired places.

Shaked et al.104) reported high-frequency GT in plants expressing the yeast DNA repair protein, Rad54 (GenBank accession no. NP_011352). Rad54 is a member of the Swi2/Snf2 family of ATP-dependent chromatin-remodeling factors,105) which play diverse roles in HR. In addition to nucleosome redistribution, these roles include interaction with the Rad51 protein, stabilization of the Rad51-single-strand DNA nucleofilament, joint-molecule formation, and post-synaptic steps.106–108) Rad51, the eukaryotic homolog of bacterial RecA recombinase, plays a central role in HR.109–114) The importance of Rad54-like activity for GT in various species115) suggests that Rad54-like proteins might be rate-limiting factors in GT in plants.
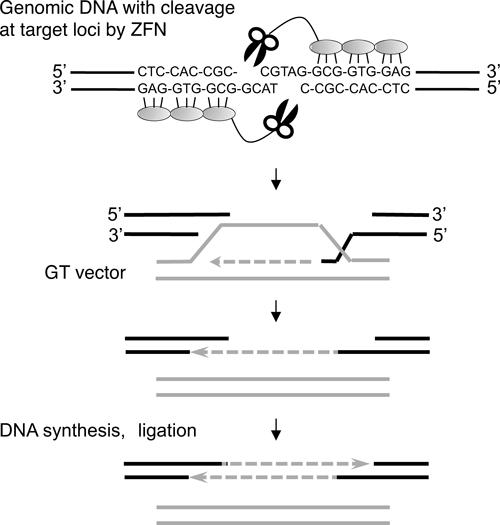
HR is a basic mechanism used to repair DNA damage, more precisely, DNA double-strand breaks (DSBs). Indeed, induction of DSBs at specific genomic locations has been shown to stimulate HR in a wide range of animal and plant species (for a review, see116–118)). For example, expression of HO and I-SceI endonucleases enhanced intrachromosomal HR in tobacco and Arabidopsis plants.119–121) Furthermore, expression of I-SceI has been shown to increase extrachromosomal HR in plants by up to 100-fold.122) Nevertheless, the limited number of naturally occurring rare-cutting endonucleases and the difficulties involved in reengineering these enzymes for novel DNA-target specificities123) greatly limit their use for GT experiments in plants. In contrast, zinc finger nucleases (ZFNs) could represent a viable alternative to rare-cutting nucleases for genome editing in plant cells. ZFNs are synthetic restriction enzymes that can be designed specifically to cleave virtually any long stretch of double-stranded DNA sequences.124–126) A ZFN monomer consists of two domains: an artificially prepared zinc finger domain and a non-specific DNA cleavage domain from the FokI DNA restriction enzyme (Fig. 6). Dimerization of the FokI domain is crucial for its enzymatic activity. Thus, digestion of target DNA can be achieved when two ZFN monomers bind to their respective DNA target configuration. ZFN-induced DSBs enhance GT at engineered loci in human cells127) and in plants.46,128–130) Shukla et al.130) and Townsend et al.46) describe GT modification of the maize gene encoding inositol-1,3,4,5,6-pentakisphosphate 2-kinase and the tobacco ALS gene, respectively. Because the zinc-finger domain131,132) can be engineered to target novel DNA sequences,133,134) ZFNs have been exploited widely in eukaryotic systems for the engineering of endogenous genome loci (for a review, see135)).
Recently, a novel DNA-binding domain in a protein family called transcription activator-like effectors (TALEs) was described.136,137) Several groups have shown that TALENs (TALEs fused with the catalytic domain of the FokI nuclease) can create targeted DSBs in vivo for genome editing.138–141) In ZFNs, one DNA-binding motif recognizes three nucleotides, but, in TALENs, one DNA-binding domain can recognize one nucleotide in the target DNA sequence.136,137) Thus, TALENs are more flexible.
Many agronomically valuable traits and natural variations are derived from a small number of point mutations. Among such phenotypes, herbicide tolerance is one of the most feasible for GT because the tolerant trait itself can be used as a selection marker for GT events. Recently, the crystal structures of herbicides in complex with their protein targets have become available,50,142) and such information offers explanations as to how plants have developed herbicide tolerance and will aid in the design of novel herbicides and herbicide-tolerant crops. A combination of techniques incorporating structural chemistry, transgenic approaches, conventional breeding, and GT promises to improve weed control in agro-ecosystems.
The manner in which herbicide-tolerant crops are created affects the process and cost of obtaining permission for commercialization. To avoid the risks associated with GM crops, many new breeding and genetic modification techniques, such as site-directed mutagenesis and the targeted deletion or insertion using ZFNs or TALENs, have been developed. Indeed, the US Department of Agriculture (USDA) ruled that, due to the lack of genetic components from plant pests, herbicide-tolerant lawn grass that had been transformed with a herbicide-tolerance type of EPSPS gene taken from other plants by using a gene gun fell outside of its regulatory authority.143) As more novel transgenic approaches and useful plants are developed, the regulation of such plants will receive more attention.
We thank T. Shimizu for supplying ALS-inhibiting herbicide and for contributing to the gene targeting experiments of rice ALS. This work was supported by the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation, GMC-0001). This work was also supported by a PROBRAIN (Program for Promotion of Basic Research Activities for Innovative Biosciences) grant to S.T. from the Bio-Oriented Technology Research Advancement Institution (BRAIN) of Japan. A part of this study was also supported financially by the Budget for Nuclear Research of the Ministry of Education, Culture, Sports, Science and Technology, based on screening and counseling by the Atomic Energy Commission. M. Endo was supported by a Grant-in-Aid for JSPS (Japan Society for the Promotion of Science) fellowship.