2021 年 46 巻 3 号 p. 283-289
2021 年 46 巻 3 号 p. 283-289
Meta-diamide insecticides including broflanilide have a high insecticidal activity by acting on RDL GABA receptors. Both membrane potential assays and docking studies suggest that the target site of meta-diamides is different from that of conventional noncompetitive inhibitors, such as fipronil. In fact, meta-diamides are effective against cyclodiene- and fipronil-resistant pests that carry target-site mutations. Dinotefuran uniquely possesses a tetrahydrofuran ring, whereas other neonicotinoids possess aromatic rings. Moreover, dinotefuran has been reported to be effective against imidacloprid-resistant strains. A docking study predicted the weak binding of dinotefuran to cytochrome P450s which are associated with imidacloprid resistance. Metabolic assays revealed that dinotefuran was not metabolized by these cytochrome P450s. These findings suggest that the lack of metabolic activity of P450s against dinotefuran causes a low level of cross-resistance.
Pesticides are an essential component of food security. They help farmers grow more food on less land by protecting crops from pests, diseases, and weeds. However, the effectiveness of pesticides is threatened by the evolution of resistant strains. There are various resistance mechanisms, such as target site mutations and metabolic breakdown. The elucidation of pesticide target-site and resistant mechanisms provides fundamental knowledge about the genetics, biochemistry, and physiology of target species. In turn, these insights offer greater prospects to develop or fine-tune strategies to minimize the impact of resistance on pest management.1) Therefore, pesticide manufacturers must undertake extensive research to understand the action and resistance mechanism of their pesticides.
With a rapidly growing field of molecular biology and structural biology, computational science becomes a promising rational approach in the elucidation of pesticide target-site and resistant mechanisms as well as pesticide design.2) In addition, theoretical models using computational science have helped understand their action mechanisms.3,4)
In this study, a computational method was applied to elucidate the mode of actions of pesticides and their resistant mechanisms, focusing on meta-diamide insecticides and dinotefuran (DTF).
Broflanilide (Fig. 1), which was discovered by Mitsui Chemicals Agro Inc., has a unique chemical structure characterized as a meta-diamide, and it has a high activity against various pests, including Coleopteran and Thysanopteran pests.5) The insect ionotropic γ-aminobutyric acid (GABA) receptor is a ligand-gated chloride channel and an important target of insecticides, such as cyclodienes and fipronil. Cyclodienes and fipronil are noncompetitive antagonists (NCAs) of the GABA receptor and are classified as Insecticide Resistance Action Committee (IRAC) group 2 chemicals. Meta-diamides induce excitatory symptoms, such as convulsions and paralysis.5) Similar symptoms were observed with NCAs. A meta-diamide, 3-benzamido-N-(2-bromo-4-(perfluoropropan-2-yl)-6-(trifluoromethyl) phenyl)-2-fluorobenzamide (meta-diamide 7, Fig. 1), had a higher larvicidal activity against common cutworm Spodoptera litura than that of fipronil.6) Meta-diamide 7 also had a higher inhibitory activity against the RDL GABA receptors of S. litura than that of fipronil.6) Meta-diamide derivatives show a linear relationship (R2=0.94) between their larvicidal activity and RDL GABA receptor inhibitory activity against S. litura, suggesting that the GABA receptor is the toxicologically relevant target of meta-diamides.6)
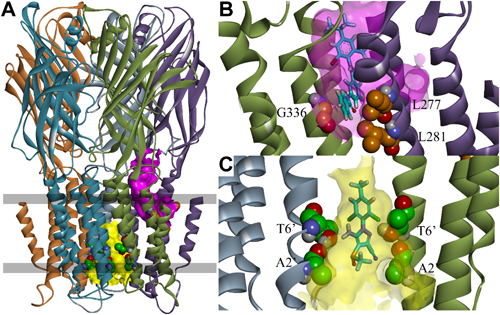
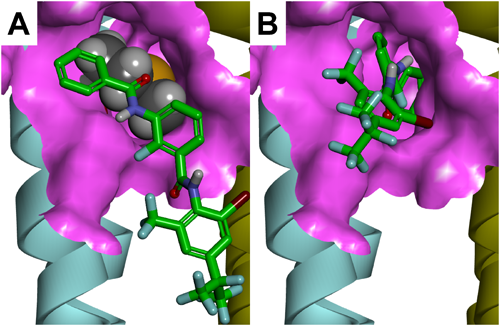
GABA receptors have five subunits; each subunit contains a large extracellular domain and four membrane-spanning regions designated M1–M4. A homology model of Drosophila melanogaster RDL GABA receptors (DM-RDL) was constructed to identify the binding site of meta-diamides.6) The numbering of the amino acid sequence of DM-RDL subunits is described in Fig. 4 in a study by Ffrench-Constant et al.7) NCAs are known to act on pores formed by M2s (Fig. 2A, C).6) A docking study suggested that meta-diamide 7 could act on a intersubunit pocket near the glycine residue 336 (G336) in the M3 of DM-RDL receptors (Fig. 2A, B).6)
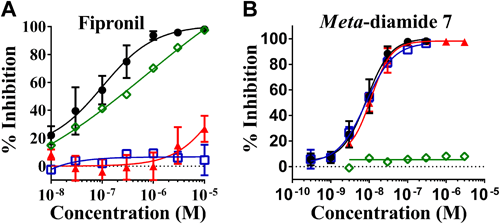
Using a membrane potential assay, the effects of meta-diamide 7 and fipronil on mutant DM-RDLs were examined. Fipronil had a little or no inhibitory activity against both A2′N and A2′S·T6′S mutant receptors, which were reported to confer resistance to NCAs (Fig. 3A). In contrast, meta-diamide 7 inhibited these mutant receptors at the same level with wild-type receptors (Fig. 3B). Although G336M mutation had a little effect on the inhibitory activities of fipronil, the G336M mutation abolished the inhibitory activities of meta-diamide 7 (Fig. 3). Both the docking studies and the membrane potential assays suggested that the binding site of meta-diamides was different from that of NCAs.6,8)
Pests have gained widespread resistance to NCAs via A2′S, A2′G, and A2′N mutations in the M2 region of GABA receptors.9–11) The predicted different binding sites between meta-diamides and NCAs suggested that meta-diamides will not show cross-resistance to biotypes carrying the A2′ mutations. In fact, no cross-resistance between meta-diamides and NCAs was observed.12) The resistant strains, which carry the A2′ mutations in the GABA receptor, were resistant to dieldrin and fipronil. In contrast, both meta-diamide 7 and broflanilide were effective against the resistant strains.12) Based on these findings, broflanilide was classified into a new IRAC group 30, GABA-gated chloride channel allosteric modulators.
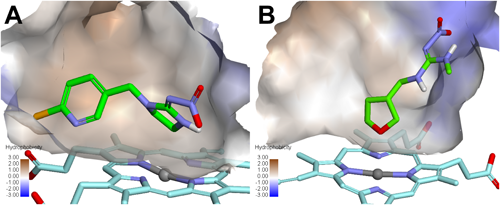
1.3. Selectivity of meta-diamides with mammal GABA receptorsAlthough meta-diamides inhibited the DM-RDL receptor with a high potency, low-level inhibitory activities of meta-diamides have been demonstrated against the human GABA type A receptor (GABAAR) α1β2γ2S and β3, mammalian GABAAR α1β3γ2S, and human glycine receptor (GlyR) α1 and α1β.13,14) The G336 in the DM-RDL subunit is essential for the high inhibitory activity of meta-diamide 7.6) The alanine residue 288 in human GlyR α1 and methionine residue 286 (M286) in human GABAAR β3 are the equivalent positions of G336 in DM-RDL. The equivalent glycine mutations A288G and M286G dramatically increased the inhibitory activities of meta-diamide 7, indicating that the glycine residue in M3 is important for the binding of meta-diamides to mammalian and insect receptors (Fig. 4).14) A homology model of human GABAAR β3 homomers in a closed state was made using human GABAAR α1β3γ2 (PDB entry code 6HUK) as a template. As shown in Fig. 5, the M286 in human GABAAR β3 subunit is positioned at the entrance of the intersubunit pocket, and the bulky side chain of M286 reduced the size of the entrance. Based on the docking studies of meta-diamide 7 to human GABAAR β3 homomers using the CDOCKER module of BIOVIA Discovery Studio 2019, the interaction energies of meta-diamide 7 in top-scoring poses were −42.0 kcal mol−1 for the M286G mutant and −27.4 kcal mol−1 for the wild type, indicating that it was more favorable for meta-diamide 7 to bind to the M286G mutant than to the wild type.14) Meta-diamide 7 was attached to the surface of the wild-type receptor and was not allowed to interact with the intersubunit pocket, whereas most of its parts were inserted into the pocket of the M286G mutant receptor (Fig. 5). These results suggest that the M286G mutation of human GABAAR is important to obtain a favorable interaction between meta-diamide 7 and the intersubunit pocket. The amino acids of human GABAARs at positions equivalent to the G336 of DM-RDL are not glycine residues, except for GABAAR π, whose amino acids around the intersubunit pocket had a low similarity with those of insect GABA receptors.14) Thus, meta-diamides are expected to be highly specific for insect GABA receptors.
Imidacloprid (IMI) showed an excellent efficacy against globally important crop pests, such as the peach–potato aphid Myzus persicae, whitefly Bemisia tabaci, and brown plant hopper Nilaparvata lugens (Stål).15) However, the intensive use of IMI resulted in the development of IMI resistance.15) The most common mechanism of IMI resistance is the overexpression of cytochrome P450 monooxygenase enzymes (CYPs). The overexpression of CYP6CM1 and CYP6ER1 confers IMI resistance to B. tabaci and N. lungens, respectively.16–18) In M. persicae, both CYP6CY3 overexpression and target-site mutation (R81T) confer IMI resistance.19–22) Although these pests showed cross-resistance to other neonicotinoids, DTF has been reported to be effective against IMI-resistant strains.23–28)
2.2. Binding affinity of DTF to cytochrome P450s that confer IMI resistanceHomology models of CYP6CM1 and CYP6ER1 were built based on human CYP3A4 (PDB entry code 4D75).29) Glide was used to estimate the binding poses and scores of IMI and DTF for the active site of these CYPs. As shown in Fig. 6, the active sites of these CYSs are hydrophobic pockets, suggesting that hydrophobic compounds are preferred for binding. The Glide scores of IMI and DTF are −4.9 and −3.5 for CYP6CM1 and −5.4 and −3.5 for CYP6ER1, respectively. According to the Glide scores, the binding of DTF to these CYPs was weaker than that of IMI, which is consistent with the previous calculations on CYP6CM1.30) To confirm this prediction experimentally, a competition between a substrate and IMI or DTF for CYP6CM1 was evaluated using Luciferin-MultiCYP as a nonselective CYP450 bioluminescent substrate.31) IMI at a concentration of 640 μM completely inhibited CYP6CM1-catalyzed Luciferin demethylation, whereas DTF at the same concentration only produced a weak inhibition.31) These results suggested that the differences in resistance level between IMI and DTF may be explained by the weaker binding of DTF to the CYPs associated with IMI resistance.
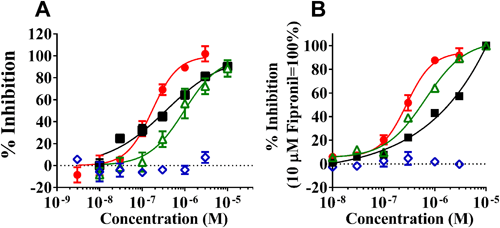
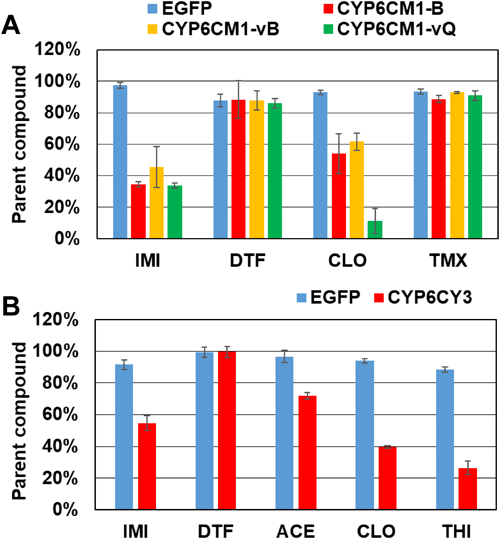
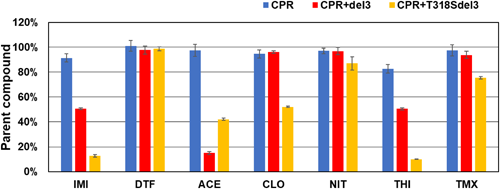
The metabolic activities of M. persicae CYP6CY3 and B. tabaci CYP6CM1 variants against DTF and other neonicotinoids (Fig. 7) were evaluated using Drosophila S2 cells stably expressing these CYPs.31,32) After 2 days of incubation in S2 cells stably expressing the CYP6CM1 variants, IMI was decreased by three CYP6CM1 variants, and hydroxyl-IMI was generated.31) Clothianidin was also metabolized by these CYP6CM1s, but DTF and thiamethoxam were not metabolized (Fig. 8A). After 4 days of incubation, CYP6CY3 showed a metabolic activity against IMI, acetamiprid, clothianidin, and thiacloprid, but it had no activity against DTF (Fig. 8B).32) The metabolism of neonicotinoids was also measured using the microsomes prepared from Sf9 cells expressing the CYP6ER1 variants.33) After 2 hr of incubation, both CYP6ER1 variants metabolized IMI, acetamiprid, and thiacloprid, but they did not metabolize DTF (Fig. 9).33) One of the CYP6ER1 variants metabolized clothianidin and thiamethoxam. Nitenpyram was metabolized slightly by one variant. Comparing the octanol-water partition coefficient (log P) shown in Fig. 9, hydrophilic compounds are less likely to be metabolized, which is consistent with human CYPs.34) These findings indicate that IMI-resistant biotypes carrying the overexpression of these CYPs will not have a strong resistance to DTF. However, recent studies, which used N. lugens selected by neonicotinoids, indicated that CYP6ER1 confers cross-resistance to most neonicotinoids, including DTF.35–38) Our results indicated that CYP variants showed a different substrate selectivity (Figs. 8, 9). Many CYP6ER1 variants have also been reported,18) and one of these variants might be at risk to confer high resistance to DTF. Therefore, it is important to determine which variant is associated with resistance to each neonicotinoid and to continuously monitor the susceptibility of field strains to each neonicotinoid.
In this study, the novel mode of action of meta-diamide insecticides was demonstrated by combining in vitro assays and computational methods. A glycine residue in M3 is important for the high selectivity of insect GABA receptors compared with those of mammals. It was also reported that the CYPs associated with IMI resistance do not metabolize DTF, probably because a highly hydrophilic DTF is a weak binder for these CYPs. Computational science is a promising rational tool to elucidate pesticide target-site and resistant mechanisms by combining the rapidly growing field of molecular biology and structural biology.
I am grateful to the Pesticide Science Society of Japan, which gave me an award. I thank the people who provided me with useful advice, support, and cooperation. I am deeply grateful to the members of Mitsui Chemicals Agro Inc. for their kind support and providing opportunities to study about the novel pesticides. Lastly, I express my sincere acknowledgement to Prof. Yoshihisa Ozoe of Shimane University and Dr. Toshifumi Nakao of Mitsui Chemicals Agro Inc. for their kind guidance, support and collaboration.