2014 Volume 64 Issue 1 Pages 23-37
2014 Volume 64 Issue 1 Pages 23-37
Most wild plants and some crops of the Brassicaceae express self-incompatibility, which is a mechanism that allows stigmas to recognize and discriminate against “self” pollen, thus preventing self-fertilization and inbreeding. Self-incompatibility in this family is controlled by a single S locus containing two multiallelic genes that encode the stigma-expressed S-locus receptor kinase and its pollen coat-localized ligand, the S-locus cysteine-rich protein. Physical interaction between receptor and ligand encoded in the same S locus activates the receptor and triggers a signaling cascade that results in inhibition of “self” pollen. Sequence information for many S-locus haplotypes in Brassica species has spurred studies of dominance relationships between S haplotypes and of S-locus structure, as well as the development of methods for S genotyping. Furthermore, molecular genetic studies have begun to identify genes that encode putative components of the self-incompatibility signaling pathway. In parallel, standard genetic analysis and QTL analysis of the poorly understood interspecific incompatibility phenomenon have been initiated to identify genes responsible for the inhibition of pollen from other species by the stigma. Herewith, we review recent studies of self-incompatibility and interspecific incompatibility, and we propose a model in which a universal pollen-inhibition pathway is shared by these two incompatibility systems.
Flowering plants have evolved a number of intraspecific and interspecific pre-fertilization pollination barriers that allow pistils to control which of the many pollen grains that arrive at the stigma can effect fertilization. The best-known intraspecific pollination barrier is self-incompatibility (De Nettancourt 2001), which is considered to have evolved as a mechanism for the avoidance of inbreeding depression and the maintenance of genetic variation in populations. For their part, interspecific incompatibility barriers prevent hybridization between different species and thus maintain the identity of each species.
The self-incompatibility response of the Brassicaceae has been well studied, especially in Brassica rapa, B. oleracea, and B. napus, which include major vegetable and oil crops, and also in the genus Arabidopsis, including the model plant A. thaliana. In this self-incompatibility system, incompatible pollen is manifested at the surface of stigma epidermal cells by the failure of pollen grains to germinate and produce pollen tubes that elongate into the epidermal cell wall (Fig. 1). Genetic studies carried out in the 1950s revealed that this trait is controlled by variants of a single locus, the S locus, (Bateman 1955). In the 1960s, immunochemical analysis of stigmas led to the identification of proteins encoded by genes at the S locus (Nasrallah and Wallace 1967a). And starting in the 1980s, molecular genetic analyses identified the S-locus genes that are responsible for the recognition of “self” pollen by the stigma. Many alleles of these genes have now been sequenced, and the sequence information has contributed to biological, physiological, biochemical, genomic, and genetic studies of self-incompatibility as well as to practical breeding utilizing this trait. Several review articles published recently have provided a general understanding of self-incompatibility in the Brassicaceae and other plant families (Iwano and Takayama 2012, Kitashiba and Nishio 2009, Rea and Nasrallah 2008, Tantikanjana and Nasrallah, 2012).
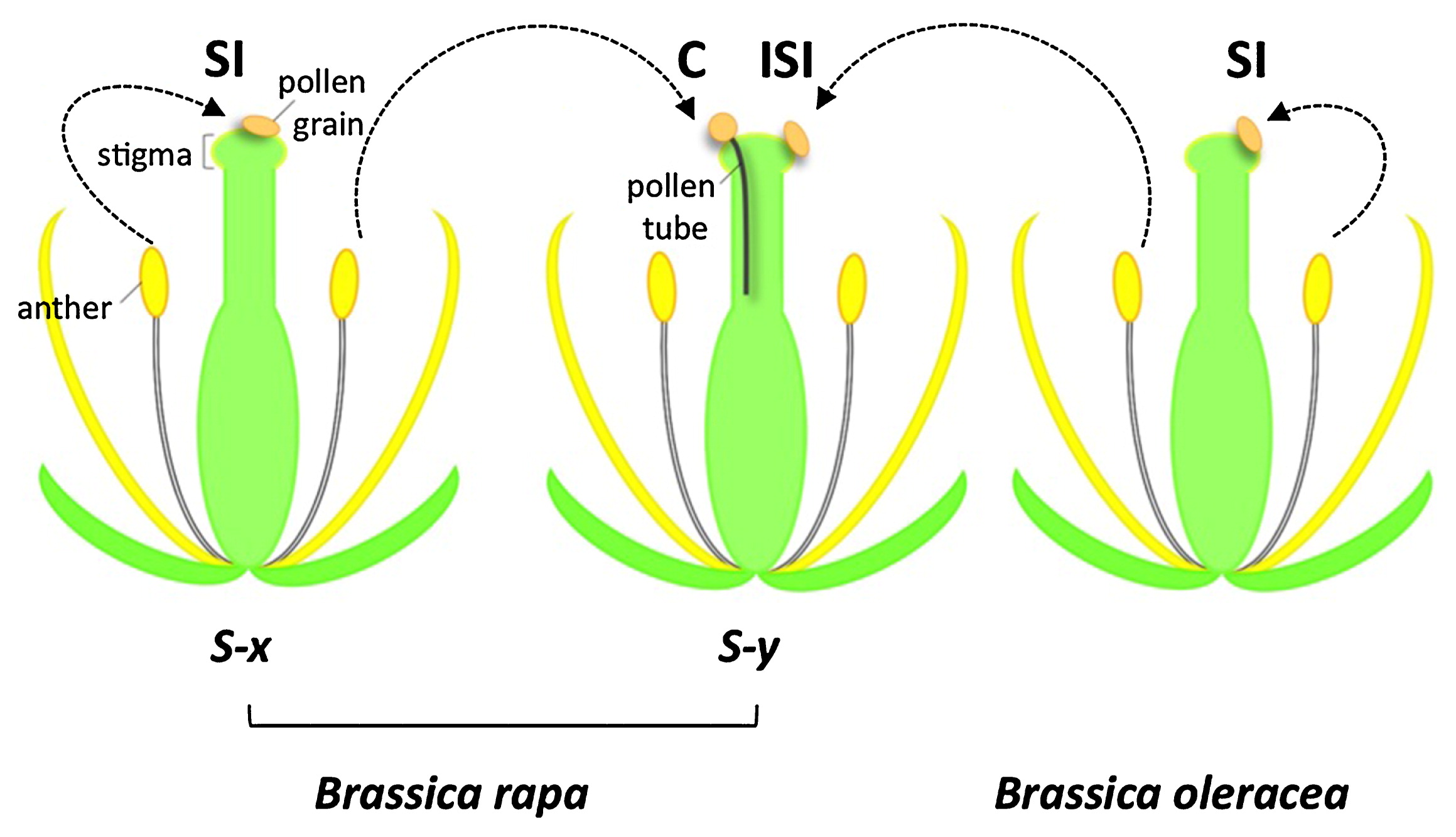
Illustration of self-incompatibility and interspecific (interspecies) incompatibility in Brassica species. SI: self-incompatibility, C: compatibility, ISI: interspecific incompatibility.
In the case of interspecific incompatibility in the Brassicaceae, cytological observations have been carried out (Hiscock and Dickinson 1993, Lewis and Crowe 1958), but the genetic basis of the phenomenon is not understood. However, because the manner in which incompatible pollen is inhibited by cells of the pistil is the same in intraspecific and interspecific pollination barriers, it has been suggested that the same or very similar signaling pathways underlie pollen recognition in the two systems. Recently, the availability of genetic maps and genomic information for an increasing number of Brassicaceae species has spurred renewed efforts to understand the genetic basis of this trait. We herewith review recent studies of self-incompatibility and interspecific incompatibility, and we propose a model of a shared universal signaling pathway that might cause inhibition of conspecific and heterospecific pollen at the stigma surface.
In the Brassicaceae, the self-incompatibility phenotype of both stigma and pollen is determined sporophytically by the diploid S-locus genotype of the parent plant and dominance relationships between S-locus variants are observed in both stigma and pollen (Thompson and Taylor 1966). Pollen inhibition at the stigma surface occurs when the pollen and stigma express the same S-locus variant. Molecular genetic studies of the Brassica S locus identified three genes, two of which were shown by functional analyses to encode proteins responsible for specific recognition of “self” pollen by the stigma. The three genes are the S-locus receptor kinase (SRK), the S-locus cysteine rich protein/S-locus protein 11 (SCR/SP11), and the S-locus glycoprotein (SLG). Because the three genes are tightly linked to each other at the S locus and their alleles are inherited as one genetic unit, the term “S haplotype” has been coined to designate variant forms of the S locus (Nasrallah and Nasrallah 1993).
Alleles of S-locus genes and S haplotypes have commonly been represented with numerical subscripts, e.g. S1, S2, etc. However, according to standard nomenclature, alleles should be shown, not with subscripts, but with “+” or “−”, e.g. S+1 or S-1 (Meinke and Koornneef 1997, Østergaard and King 2011). Hereafter, to designate S haplotypes and alleles of S-locus genes, we use “−” followed by the allele number and preceded by letters that abbreviate the species name: e.g., S haplotypes of B. rapa and B. oleracea are represented by BrS-1 and BoS-1, respectively.
Identification of S-locus genes: SLG, SRK, and SCR/SP11Of the three S-locus genes, the S-locus glycoprotein gene, SLG, was identified first. Its protein product was identified in stigma extracts by immunochemical analysis as S allele-specific antigens (Nasrallah and Wallace 1967b) and by electrophoretic analysis as having S allele-specific pI values (Nishio and Hinata 1977). In both cases, variant forms were shown to co-segregate with S alleles (Hinata and Nishio 1978, Nasrallah and Wallace 1967b). Subsequently, SLG cDNAs were cloned and sequenced (Nasrallah et al. 1985, 1987) and the amino-acid sequence of SLG was also determined by direct protein sequencing (Takayama et al. 1987). This initial sequence information enabled the determination of sequences for many additional SLG alleles in Brassica species and in Raphanus sativus (Chen and Nasrallah 1990, Kusaba et al. 1997, Sakamoto et al. 1998, Trick and Flavell 1989, ). All SLG alleles are predicted to encode a primary translational product consisting of a hydrophobic signal peptide at the N-terminus for secretion to the outside of cells, several potential N-glycosylation sites, and twelve conserved cysteine residues. The mature SLG protein is a highly polymorphic glycoprotein having many residues that vary between alleles. Moreover, the SLG gene is expressed at high levels in stigma epidermal cells as demonstrated by in situ hybridization of SLG transcripts (Nasrallah et al. 1988) and analysis of SLG promoter activity using the GUS reporter (Sato et al. 1991), and the SLG protein accumulates in the wall of stigma epidermal cells as determined by immunocytochemical analysis (Kandasamy et al. 1989).
The next S-locus gene to be identified was SRK, which was isolated by screening a genomic library with an SLG probe and was found to contain a kinase-encoding sequence and to be linked to SLG (Stein et al. 1991). The primary SRK translational product consists of an N-terminal signal sequence, an extracellular domain named “S” domain that is highly similar to SLG, followed by a transmembrane domain and a serine/threonine protein kinase domain toward the C-terminus. Like SLG, the SRK gene is predominantly expressed in stigma epidermal cells (Nasrallah et al. 1994). Also similar to SLG, the S domain of SRK is highly polymorphic, with some variants exhibiting more than 30% amino-acid sequence divergence. Comparison of nucleotide sequences and deduced amino-acid sequences demonstrated that the SRK S domain and the SLG encoded by the same S haplotype share more than 90% amino-acid sequence identity (Hatakeyama et al. 1998b; Sato et al. 2002, Stein et al. 1991, Watanabe et al. 1994).
The last S-locus gene to be identified was the SCR/SP11 gene. In 1999, two groups, Schopfer et al. (1999) and Suzuki et al. (1999), surveyed the S locus by cloning and sequencing of the S-locus region in B. rapa, and identified a gene adjacent to SLG and SRK that exhibited anther-specific expression. Sequence analysis of this SCR/SP11 gene predicted a protein having an N-terminal signal peptide, which when cleaved, would result in a mature cysteine-rich protein that is secreted, small in size (~50 amino acids), and basic in nature. Sequence comparisons showed that the mature SCR/SP11 protein is highly polymorphic, with less than 50% amino-acid similarity shared by variants within the same species (Okamoto et al. 2004, Sato et al. 2002, Schopfer et al. 1999, 2000, Watanabe et al. 2000). Only a few amino acids are conserved in most SCR/SP11 variants: eight cysteines, a glycine between the first and second cysteines, and an aromatic amino-acid residue between the third and fourth cysteines (Schopfer et al. 1999, 2000, Takayama et al. 2000, Watanabe et al. 2000). Furthermore, GUS reporter analysis of the SCR/SP11 promoter (Schopfer et al. 2000) and in situ hybridization of SCR/SP11 transcripts (Takayama et al. 2000, Kusaba et al. 2002) demonstrated that the gene was predominantly expressed in the anther tapetum, as expected for the sporophytic control of pollen self-incompatibility specificity in the Brassicaceae.
The S haplotypes of Brassica and Raphanus speciesA large number of S haplotypes have been identified in B. rapa and B. oleracea and in the closely-related Raphanus sativus by pollination tests, electrophoretic analysis of stigma proteins, analysis of DNA polymorphism in SLGs or SRKs, and determination of SLG, SRK, and SCR/SP11 sequences. In B. oleracea, among the 49 S haplotypes reported to date (Oikawa et al. 2011), sequences of the SLG, SRK or SCR/SP11 alleles have been determined for all S haplotypes with the exception of four S haplotypes, S-10, S-56, S-66 and S-67. In B. rapa, the sequences of the three genes have been determined for 44 S haplotypes (Oikawa et al. 2011). On the basis of the extent of sequence similarity shared by SLG and SRK alleles, S haplotypes have been grouped into two major classes, designated class-I and class-II S haplotypes (Nasrallah et al. 1991). Because of the availability of these sequences, S haplotypes have been standardized in B. oleracea and B. rapa, and collections of S haplotypes have been established in the two species. However, it is likely that additional S haplotypes exist which have not been characterized. Nou et al. (1993) used calculations based on the number of S haplotypes in wild populations in Japan and Turkey to estimate that more than 100 S haplotypes exist in B. rapa. Ockendon (2000) speculated that the total number of S haplotypes in B. oleracea is approximately 50 based on his long-time study of S-haplotype collections.
In contrast to the situation in Brassica, some confusion exists in the numbering of S haplotypes in R. sativus. Sakamoto et al. (1998) and Okamoto et al. (2004) reported on 20 R. sativus S haplotypes (S-1 to S-19 and S-21) along with the sequences of their SLG, SRK, and/or SCR/SP11 alleles. Lim et al. (2002) also reported the sequences of these genes for 10 S haplotypes that they named S-1 to S-10, but these S haplotype numbers do not refer to the same numbers used by Sakamoto et al. (1998) and Okamoto et al. (2004). Additionally, Niikura and Matsuura (1999) used pollination assays and PCR-RFLP analyses to identify 37 S haplotypes from Japanese, Asian, and European domesticated and commercial cultivars, and numbered them S-201 to S-237. Since the latter S haplotypes are not associated with sequence information for the SLG, SRK and SCR/SP11 alleles, it is not known if they are identical to any of the S haplotypes identified in the studies described above. Exchanges of plant materials between researchers and breeders and the establishment of a unified nomenclature of S haplotypes are necessary to avoid confusion regarding the identity of S haplotypes in R. sativus.
Owing to the extensive S haplotype collections in Brassica species, close relationships between SLGs and the S-domains of SRKs have been unveiled. The high degree of identity shared by the two sequences suggests the occurrence of gene conversion between SLG and the S domain of SRK in the same S haplotype. This conclusion is supported by analysis of the progeny of a B. rapa commercial cultivar showing self-compatibility, in which part of the S domain of SRK was replaced with the corresponding part of SLG from the same S haplotype (Fujimoto et al. 2006c). The occurrence of such gene conversion events has important implications. If a mutation causes an amino-acid substitution within the SRK S domain, which is the domain that perceives SCR (see below), the recognition specificity of SRK will be changed, causing a switch to self-compatibility. The resulting self-compatible plant would suffer inbreeding depression produced by repeated self-pollinations and is expected to become extinct. Therefore, to maintain recognition specificity, the mutation in SRK must be amended. Gene conversion between SRK and SLG is thought to provide a mechanism for the amendment of the mutated segment of the SRK S-domain by replacement with the corresponding non-mutated segment from SLG (Sato et al. 2002).
Comparison of nucleotide and amino-acid sequences of the SLG, SRK, and SCR/SP11 alleles also contributed to the identification of interspecific pairs of S haplotypes, i.e. pairs of S haplotypes from different species, whose SLG, SRK, and SCR/SP11 sequences are more similar to each other than to those derived from other S haplotypes within the same species (Kimura et al. 2002, Kusaba et al. 1997). So far, eighteen interspecific pairs were found in comparisons of S haplotypes from B. rapa and B. oleracea (Kimura et al. 2002, Kusaba et al. 1997, Kusaba and Nishio 1999, Oikawa et al. 2011, Sato et al. 2003) and one intergeneric pair was identified by comparing B. rapa and R. sativus sequences (Okamoto et al. 2004, Sato et al. 2004). The use of several methods, including interspecific hybridization between B. rapa and B. oleracea (Kimura et al. 2002), transformation of B. rapa with B. oleracea or R. sativus SCR/SP11 alleles, and bioassays of SCR/SP11 proteins synthesized in bacteria (Sato et al. 2003, 2004, 2006) have revealed that the interspecific pairs of S haplotypes retain the same recognition specificities. These reports suggest that each interspecific and intergeneric pair of S haplotypes originated from a common ancestral S haplotype, and that the diversification of S haplotypes occurred before speciation. Further insight into the diversification of S haplotypes will be obtained by analysis of S-haplotype collections in self-incompatible species of other genera of the Brassicaceae.
As the number of characterized S haplotypes increases, the development of methods for the identification of S haplotypes becomes increasingly important. Although classical methods, such as pollination tests and isozyme analysis, allow discrimination between S haplotypes, a method for analysis of DNA polymorphisms by PCR-RFLP, was developed based on polymorphisms in SLG and SRK alleles (Brace et al. 1993, Nishio et al. 1994). Because this method is simple and reliable, it is used for F1 hybrid breeding and seed purity tests as well as for basic research on self-incompatibility. However, the PCR-RFLP method cannot easily identify the S genotypes of heterozygous plants because of complicated band patterns and it is not applicable to the few S haplotypes that lack SLG (Sato et al. 2002). To replace the PCR-RFLP method, Fujimoto and Nishio (2003) developed a dot-blot hybridization method that utilizes the highly variable sequences of SCR/SP11 alleles. More recently, this method was improved to be more reliable and simpler (Takuno et al. 2010). It now enables allele-specific detection of SCR/SP11 with high signal intensities and it can be used to identify most known S haplotypes of B. rapa (40 S haplotypes) and 33 S haplotypes of B. oleracea (Oikawa et al. 2011). Owing to these efforts, a classification system for S haplotypes and associated S tester lines is now available for B. rapa and B. oleracea (Oikawa et al. 2011).
Genomic structure of the Brassica S locusAnalysis of genomic organization in a small number of S haplotypes has shown that the physical size of the S-locus core region, i.e. the region containing the SLG, SRK, and SCR/SP11 genes, tends to be shorter and less variable in B. rapa than in B. oleracea (Boyes and Nasrallah 1993, Suzuki et al. 2000a), due at least in part to the insertion of several retrotransposon-like sequences in B. oleracea S haplotypes (Fujimoto et al. 2006a, Kimura et al. 2002). However, further comparative genomic studies of a much larger number of S haplotypes is required to determine if these differences reflect true differences between species or simply variation among different S haplotypes within a species. In any case, sequence analysis of the S-locus region identified several genes of unknown function in regions flanking the S-core region, two of which, the SP6 (S-locus protein 6) and the SLL2 (S-locus-linked 2) genes, delimit the ends of the S haplotype.
Comparative analysis of class-I S haplotypes in B. rapa, B. oleracea, and B. napus (Boyes et al. 1997, Cui et al. 1999, Shiba et al. 2003) have shown that the genomic regions outside the S-locus core exhibit a high degree of synteny and sequence similarity between different S haplotypes, whereas the S-locus core region is highly polymorphic and enriched in S-haplotype-specific intergenic sequences. In both B. rapa and B. oleracea, the organization of the S locus differs substantially between class-I and class-II S haplotypes. In the class-I BrS-8, BrS-9, BrS-46, BrS-47 and BrS-54 haplotypes, the order of genes is SLG–SCR/SP11–SRK (Fujimoto et al. 2006a, 2006b, Suzuki et al. 1999, Takuno et al. 2007). In contrast, in the class-II BrS-60 haplotype, the order of SRK and SCR/SP11 was the reverse of that in class-I S haplotypes (Fukai et al. 2003). Furthermore, the direction of transcription of the SRK, SCR/SP11, and SLG genes in four B. rapa class-II S haplotypes is completely conserved, although the region between SRK and SCR/SP11 is highly diverse (Kakizaki 2006).
The diversity observed in the structure and sequence of different S haplotypes within the same species is considered to contribute to the suppression of recombination in the S-locus region (Boyes et al. 1997). Such suppression of recombination is thought to be required for maintaining the linkage of matched alleles of the S-locus recognition genes and consequently for persistence of the self-incompatibility trait, because recombination events that disrupt the recognition gene complex are expected to produce non-functional recombinant S haplotypes. Plants harboring these non-functional S haplotypes are expected to exhibit increased homozygosity for deleterious mutations and eventual elimination of the recombinant S haplotype due to inbreeding depression (Boyes et al. 1997, Casselman et al. 2000).
However, a genetic analysis of the frequency of recombination across the B. rapa S locus did not show a reduced rate of crossovers within the S-locus region relative to flanking regions or other regions of the genome (Casselman et al. 2000). Moreover, although no recombination events have apparently involved SCR/SP11 and the S-domain of SRK, the kinase domain of SRK and other S locus-linked genes not responsible for self-recognition have experienced recombination between S haplotypes (Takuno et al. 2007). Further studies are required to determine if recombination is actually suppressed in the S-locus region and if estimates of recombination frequencies will differ depending on which S haplotypes or combinations of S haplotypes are analyzed.
S haplotypes in other self-incompatible species of the Brassicaceae: Arabidopsis speciesSubsequent to the identification of S-locus genes in Brassica, orthologs of these genes were sought in other self-incompatible species of the Brassicaceae, especially Arabidopsis lyrata, and to a lesser extent A. halleri, Capsella grandiflora, and Leavenworthia (Chantha et al. 2013). The S locus and its genes have been particularly well characterized in the genus Arabidopsis, which is considered to be closer to ancestral species of the Brassicaceae than Brassica. To date, many complete or partial sequences of SRK and SCR alleles have been obtained from A. lyrata and A. halleri (Bechsgaard et al. 2006, Boggs et al. 2009a, Kusaba et al. 2001, Prigoda et al. 2005, Schierup et al. 2001, Tsuchimatsu et al. 2010). In addition, the basic features of the Arabidopsis S locus were described, first by structural and transcriptional analysis of two A. lyrata S haplotypes (Kusaba et al. 2001) and subsequently by sequence analysis of additional A. lyrata S haplotypes (Goubet et al. 2012, Guo et al. 2011).
These studies showed that, similar to Brassica S haplotypes, A. lyrata S haplotypes differ in the order and orientation of the S-locus recognition genes, as well as in the physical distances between these genes owing to the insertion of variable numbers and types of transposable elements (Goubet et al. 2012, Guo et al. 2011, Kusaba et al. 2001). However, the S locus of Arabidopsis species, as well as that of Capsella species (Nasrallah et al. 2007), differs from the Brassica and Raphanus S locus in two major features: it lacks an SLG gene and it is not flanked by homologues of the SP6 and SLL2 genes as in Brassica, but is rather delineated by the ARK3 gene on one end and the U-box gene PUB8 on the other end (Conner et al. 1998, Goubet et al. 2012, Guo et al. 2011, Kusaba et al. 2001). Thus, the S locus occupies a region of the Arabidopsis and Capsella genomes that is not syntenic with the S-locus region in Brassica, suggesting that a translocation of the locus occurred in Brassica, likely as a consequence of the extensive genomic rearrangements that are known to have occurred in this genus (Kusaba et al. 2001).
The expression of the SLG and SRK genes in stigma epidermal cells and of the SCR/SP11 gene in the tapetum is consistent with a role for the products of these genes as determinants of self-incompatibility specificity in stigma and pollen, respectively. Proof that genes actually fulfill these functions was obtained by transformation experiments. Thus, the SCR/SP11 gene was shown to encode the pollen determinant of self-incompatibility specificity by Schopfer et al. (1999) and subsequently by Shiba et al. (2001): in both studies, pollen grains from plants transformed with an allele of this gene was shown to be specifically inhibited on stigmas expressing the corresponding SRK variant.
For SLG and SRK, determining a role in self-incompatibility was less straightforward. The self-incompatibility response is developmentally regulated as a function of stigma development, such that immature stigmas are self-compatible and the ability of the stigma to inhibit “self” pollen is first observed one day prior to anthesis. Both SLG and SRK transcripts are developmentally regulated, attaining maximal levels in the stigmas of flower buds just before anthesis, coincident with the onset of the self-incompatibility response (Delorme et al. 1995, Stein et al. 1991, Watanabe et al. 1994). This expression pattern, together with the high degree of polymorphism exhibited by both SLG and SRK, raised a question about which gene functioned as the stigma determinant of specificity in the self-incompatibility response. Initial attempts to modify self-incompatibility specificity by transformation with SRK genes were not successful due to transgene-mediated silencing, which caused suppression of the SRK transgene as well as the endogenous SLG gene (Conner et al. 1997). Subsequently, Takasaki et al. (2000) found that B. rapa plants that were transformed with SRK-9 (equivalent to SRK-28) exhibited rejection of pollen grains from an S-9 homozygote, while transgenic plants harboring the SLG-9 transgene did not. In support of these results, several Brassica plants either having defects in SLG-coding sequences, e.g., BoS-18 and BoS-60, or lacking an SLG gene, e.g., BrS-32 and BrS-36, were found to exhibit a strong self-incompatibility response (Sato et al. 2002, Suzuki et al. 2000b). Furthermore, as noted earlier, the S locus of Arabidopsis and Capsella species lacks an SLG gene. These observations indicate that SRK is the sole female determinant of S-haplotype specificity in the self-incompatible response.
What then is the role of SLG? Transgenic B. rapa plants harboring both SRK-9 and SLG-9 transgenes exhibited a significantly enhanced self-incompatibility response compared with transgenic plants harboring the SRK-9 transgene alone (Takasaki et al. 2000). However, no such enhancement of self-incompatibility was found in another transgenic experiment that used SRK-910 and SLG-910 in self-incompatible Brassica napus (Silva et al. 2001). Taken together, these observations indicate that although SLG is not necessary for the self-incompatibility response, it may enhance the SRK-mediated self-incompatibility response in some S haplotypes such as BrS-9, probably because of the high sequence similarity (98%) shared by SLG and the S domain of SRK in this haplotype. A possible molecular explanation for this enhancing effect of SLG was provided by Dixit et al. (2000). Two mutant self-compatible B. oleracea strains that expressed low levels of SLG were found to accumulate normal levels of SRK mRNA, yet failed to produce SRK protein. In addition, the B. oleracea BoSRK-6 variant was found to form aberrant high molecular mass aggregates when expressed alone in tobacco leaves but not when co-expressed with BoSLG-6. These results suggest that for at least some SRK variants, SLG is important for the stabilization and proper processing of SRK and its accumulation to physiologically relevant levels in stigmas.
The fact that SRK and SCR are the sole determinants of self-incompatibility specificity was dramatically demonstrated by transfer of the self-incompatibility trait to A. thaliana, the self-compatible close relative of A. lyrata and A. halleri. A. thaliana harbors non-functional S haplotypes, designated pseudo-S ( ΨS) haplotypes that contain non-functional alleles of SRK (ΨSRK) and/or SCR (ΨSCR) that either encode truncated proteins or are highly decayed and exhibit deletions or rearrangements (Boggs et al. 2009b, Dwyer et al. 2013, Kusaba et al. 2001, Sherman-Broyles et al. 2007, Shimizu et al. 2008, Tang et al. 2007).
Transformation with functional SRK and SCR transgenes isolated from A. lyrata or C. grandiflora was sufficient to confer a self-incompatibility phenotype in several accessions of A. thaliana (Boggs et al. 2009b, Liu et al. 2007, Nasrallah et al. 2002, 2004). Furthermore, an A. thaliana accession harboring a functional SRK allele and a non-functional SCR allele exhibited self-incompatibility when transformed with the corresponding functional SCR allele from A. halleri (Tsuchimatsu et al. 2010). These transgenic complementation experiments confirm that SRK and SCR alone determine self-incompatibility specificity. Moreover, because the self-incompatibility response exhibited by A. thaliana SRK and SCR transformants was as intense as that observed in naturally self-incompatible plants, it may be concluded that the SRK-mediated signal transduction pathway is active even in this self-compatible species. As described later, the availability of transgenic self-incompatible A. thaliana plants has allowed researchers to exploit the extensive genetic resources and ease of transformation of the A. thaliana model species for analysis of the recognition and response phases of self-incompatibility.
The basis of specificity in the self-incompatibility response: allele-specific interactions between SRK and SCRSeveral studies have demonstrated that SCR/SP11 is the ligand for SRK. Takayama et al. (2001) and Kachroo et al. (2001) reported that SRK and SCR/SP11 proteins interacted only if they were derived from the same S haplotype. SRK tends to form oligomers, especially homo-oligomers, in unpollinated stigmas, i.e. in the absence of its SCR/SP11 ligand (Giranton et al. 2000, Naithani et al. 2007, Shimosato et al. 2007). Because SRK dimerization is critical for high-affinity binding of SCR/SP11 (Shimosato et al. 2007), the constitutive homo-oligomerization of SRK might serve to prime SRK for rapid activation upon ligand binding. In a search for amino-acid residues or domains that determine SRK specificity, four hypervariable (HV) regions, HV1, HV2, HV3 and the C-terminal variable region (CVR) were identified (Kusaba and Nishio 1999, Nishio and Kusaba 2000, Sato et al. 2002). Because many of these polymorphic residues have a high probability for being under positive selection (Sainudiin et al. 2005), they are surmised to be important for specificity. However, transgenic experiments in A. thaliana involving domain swapping between two pairs of A. lyrata SRK alleles and in vitro mutagenesis determined that only a very small subset of these polymorphic amino-acid residues located in two clusters within the HV1 and HV2 regions were necessary for specific recognition of self SCR/SP11 ligand and activation of the self-incompatibility response (Boggs et al. 2009c). Whether these results apply to all SRKs is not known and a general rule for the key amino acids responsible for self-recognition has yet to be clarified.
Two approaches were used in an attempt to find a key motif for self-recognition specificity in the SCR/SP11 protein. One approach involved comparative analysis of SCR/SP11 sequences from interspecific pairs of S haplotypes (Kusaba et al. 1997, Kimura et al. 2002). Based on this analysis, six regions (Regions I to VI), each delimited by conserved cysteine residues, were assigned to SCR/SP11, and domain-swapping experiments revealed that Regions III and V are necessary for recognition of “self” SRK (Sato et al. 2004). A second approach to define the residues responsible for recognition specificity in SCR was implemented by Chookajorn et al. (2004). In this study, the B. oleracea SCR-6 and SCR-13 alleles were analyzed by domain swapping of regions between the conserved cysteines and by site-directed mutagenesis to generate chimeras and mutant forms of SCR/SP11. These engineered variants were tested by ELISA and pull-down assays for binding to the extracellular S domains of SRK-6 and SRK-13 and by pollination bioassays for their ability to elicit self-incompatibility on the stigmas of S-6 and S-13 homozygotes. Surprisingly, this study showed that SCR-13 specificity was determined by only four contiguous amino-acid residues in the region between the fifth and sixth conserved cysteines, which corresponds to Region V defined by Sato et al. (2004). However, the corresponding region in SCR-6 did not effect a change in specificity when inserted into the SCR-13 backbone. Thus, different regions or combinations of regions can determine specificity in different SCR/SP11s. Consequently, it has not been possible to infer general rules for defining residues that determine SCR/SP11 specificity.
In their analysis of genetic dominance relationships between B. oleracea and B. rapa S-locus variants, Thompson and Taylor (1966) and Hatakeyama et al. (1998a) found that class-I S haplotypes are generally dominant to class-II S haplotypes in pollen. Molecular analysis in Brassica and in A. lyrata demonstrated that plants heterozygous for a dominant and a recessive SCR/SP11 allele failed to accumulate transcripts derived from the recessive SCR/SP11 allele in the anther tapetum, where SCR/SP11 transcripts are normally expressed (Kusaba et al. 2002, Shiba et al. 2002). This result indicates that transcription of a recessive SCR/SP11 allele is suppressed in the presence of a dominant SCR/SP11 allele. Interestingly, Fujimoto et al. (2006b) observed that class-I S haplotypes in which the SCR/SP11 is not transcribed due to defects in promoter activity also caused suppression of recessive class-II SCR/SP11 alleles, suggesting that expression of dominant SCR/SP11 alleles is not necessary for the suppression of recessive SCR/SP11 alleles. Subsequently, Shiba et al. (2006) demonstrated that suppression of the transcription of a recessive class-II SCR/SP11 in heterozyogtes occurred epigenetically by de novo methylation of 5′ promoter sequences in tapetum cells. Further analysis of this phenomenon (Tarutani et al. 2010) identified inverted genomic sequences similar to the sequence of class-II SCR/SP11 promoters in regions flanking the dominant SLG alleles, which produced an anther-specific trans-acting small non-coding RNA (small RNA). Furthermore, a transgene of this small RNA induced methylation of the promoter of recessive class-II SCR/SP11 alleles. Taken together, these results indicate that the small RNA is a key sequence for suppression of the class-II SCR/SP11 transcription.
However, this mode of epigenetic control may not be generally applicable to all cases of S haplotype dominance in pollen, as suggested by analysis of dominant-recessive interactions among B. rapa class-II S haplotypes. Four B. rapa class-II S haplotypes (S-44, S-60, S-40 and S-29) have been reported, which constitute a linear dominance series, BrS-44 > BrS-60 > BrS-40 > BrS-29, in which BrS-44 is the most dominant and BrS-29 is the most recessive (Kakizaki et al. 2003). Plants heterozygous for combinations of these class-II S haplotypes exhibited de novo promoter methylation and suppression of the transcription of recessive SCR/SP11 alleles in anthers (Shiba et al. 2006). However, trans-acting non-coding RNA sequences having homology with the promoters of the recessive SCR/SP11 alleles were not observed in the dominant BrS-44 haplotype, and the non-coding RNA sequences in other S haplotypes were poorly transcribed (Tarutani et al. 2010). This observation suggests that the molecular mechanism that causes promoter methylation and suppression of transcription of class-II SCR/SP11 alleles is not based on the activity of an S-locus linked small RNA species. Further sequence analysis of small RNAs and unidentified sequences in the recessive class-II SCR/SP11 promoters will no doubt be informative in this regard.
As for the dominance relationships of S haplotypes in stigmas, genetic analyses have been conducted (Hatakeyama et al. 1998a, Thompson and Taylor 1966) but no molecular explanation has been put forth. Because recessiveness of S haplotypes in the stigma is not associated with lower SRK expression levels (Hatakeyama et al. 2001), it is possible that dominance relationships are determined, not by differences in relative expression levels of SRK, but by features of the SRK protein itself. In particular, the S domain rather than the kinase domain may dictate dominance relationships, as suggested by sequence analysis of SRKs derived from S haplotypes that exhibit different allelic relationships. For example, sequence analysis of BrSRK-54, which is co-dominant with BrSRK-8 and recessive to BrSRK-46, has shown that the SRK kinase domain of these three variants are highly similar (98.3 to 100 % at amino acid level), while their S domains exhibit a much higher degree of sequence divergence (77.8 to 85.3 % at amino acid level) (Takuno et al. 2007 and our unpublished data). In support of this conclusion, a yeast two-hybrid interaction analysis of SRK S domains showed a preference for homodimerization (i.e. interaction with an identical S domain derived from the same SRK variant) over heterodimerization (i.e. interaction with an S domain derived from another SRK variant) (Naithani et al. 2007). This result suggested the hypothesis that SRK dominance relationships result from differences in the propensity of some pairs of SRKs to form heterodimers that might have reduced affinity for SCRs (Naithani et al. 2007). Further genetic and biochemical studies are required to test this hypothesis and to explain the puzzling dominance relationships of SRK alleles.
The functionally diploid B. rapa, B. nigra, and B. oleracea, which have the A, B, and C genomes, respectively (U 1935), exhibit self-incompatibility. In contrast, B. napus, B. juncea, and B. carinata, which are amphidiploid species that contain the A and C genomes, the A and B genomes, and the B and C genomes, respectively, exhibit self-compatibility. Interestingly, artificially synthesized Brassica amphidiploid plants are self-incompatible (Hinata and Nishio 1980). To explain this discrepancy, 45 lines of B. napus were analyzed by cloning of SLG, SRK, and SCR/SP11 genes. Seven S haplotypes were identified, among which BnS-1 to BnS-5 are class-I haplotypes while BnS-6 and BnS-7 are class-II S haplotypes (Okamoto et al. 2007). Four B. napus genotypes were combinations of a dominant class-I S haplotype and a recessive class-II haplotype (Okamoto et al. 2007, Tonosaki and Nishio 2010). Further analysis revealed that S-haplotype mutations were responsible for self-compatibility in three of these genotypes. In two B. napus genotypes, dominant SRK alleles had frame-shift mutations in their coding region that knocked out SRK function (Okamoto et al. 2007). By contrast, B. napus cv. ‘Westar’, which has the pollen-dominant BnS-1 haplotype in its A genome and the pollen-recessive BnS-6 haplotype in its C genome, contains functional SRK genes but has suffered loss of SCR/SP11 function for two reasons: firstly, the dominant BnS-1 SCR/SP11 allele is not expressed due to an insertion mutation in its promoter region and secondly, the recessive BnS-6 SCR/SP11 allele is suppressed by the dominant BnS-1 SCR/SP11 allele (Okamoto et al. 2007). This case of suppression is similar to the suppression of a recessive SCR/SP11 allele by a dominant non-functional SCR/SP11 in monogenomic B. rapa described by Fujimoto et al. (2006b). In support of the conclusion that self-compatibility in ‘Westar’ is due to loss of SCR/SP11 function, ‘Westar’ plants acquired the self-incompatibility trait when transformed with a functional SCR/SP11 allele from the BrS-47, which is the likely B. rapa S haplotype progenitor of BnS-1 (Tochigi et al. 2011). Taken together, these results demonstrate that a single mutation event in the SRK or SCR/SP11 genes of a dominant S haplotype can cause self-compatibility in amphidiploid Brassica plants.
Self-compatible amphidiploids have also been described in other Brassicaceae. Arabidopsis suecica, which is an amphidiploid species derived by spontaneous hybridization between A. thaliana, a self-compatible species, and A. arenosa, a self-incompatible species, is self-compatible (Mummenhoff and Hurka 1995, O’Kane et al. 1996). In addition, artificial amphidiploids generated by crossing A. thaliana and A. lyrata also exhibited self-compatibility (Nasrallah et al. 2007) due to suppression of A. lyrata-derived SRK transcripts. Because backcrossing of the amphiploid to A. lyrata caused a reversion to self-incompatibility along with restoration of SRK expression, it is likely that an epigenetic change in the A. lyrata SRK gene or an A. thaliana-derived factor caused suppression of SRK gene expression (Nasrallah et al. 2007). Thus, based on the limited information available so far, it appears that the mechanism causing self-compatibility in amphidiploid Arabidopsis species is different from that observed in Brassica amphidiploids.
While the stigma receptor kinase SRK, and its pollen ligand SCR/SP11 have been well characterized as determinants of self-recognition, the signaling pathway triggered by ligand binding and activation of the receptor and the mechanism of rapid self-pollen rejection are poorly understood. Nevertheless, molecular genetic analyses have implicated three proteins in self-incompatibility signaling: the M-Locus Protein Kinase (MLPK), the Arm repeat-Containing protein (ARC1), and the Exo70A1 component of the exocyst complex.
Classical genetic studies indicated that self-compatibility in B. rapa cv. ‘Yellow Sarson’, an Indian oil crop, is caused by two independent loci, the S locus, which contains non-functional recognition genes and the M locus (Hinata et al. 1983). Map-based cloning of the M-locus region identified a gene encoding a membrane-anchored cytoplasmic protein kinase, which was designated MLPK (Murase 2004). The MLPK gene produces two isoforms, both of which interact with SRK (Kakita et al. 2007). A single mutation causing a nonsynonymous substitution was found in the kinase domain of the ‘Yellow Sarson’ MLPK variant, which abolished autophospholylation activity and prevented localization of the protein to the cell membrane (Murase et al. 2004). Transient expression of a wild-type MLPK allele introduced by particle bombardment into the stigma epidermal cells of mutant mlpk homozygotes conferred a self-incompatibility phenotype in these cells. This result suggests that MLPK is important for the self-incompatibility response (Kakita et al. 2007). However, a stable transformation experiment that shows complementation of the self-compatibility trait by transformation of mlpk homozygotes with the wild-type MLPK gene has not been reported.
ARC1 was isolated in a yeast two-hybrid screen as a protein that interacts with and is phosphorylated by the SRK kinase domain (Gu et al. 1998). ARC1 is expressed specifically in stigma tissues and antisense suppression of ARC1 expression in a self-incompatible B. napus strain was reported to cause partial breakdown of self-incompatibility (Stone et al. 1999). ARC1 is an E3 ubiquitin ligase and is thus considered to ubiquitinate factors required for compatible pollination and cause their degradation via the proteasome protein degradation system (Stone et al. 2003). An ARC1 ortholog was identified in a region of the A. lyrata genome that shares a high degree of synteny with the Brassica ARC1 genomic region (Kitashiba et al. 2011), and transformation of A. lyrata plants with an ARC1-RNAi construct caused reduced transcription of the endogenous ARC1 gene and partial loss of self-incompatibility (Indriolo et al. 2012). Additionally, ARC1 sequences were found to be deleted in the genomes of some self-compatible Brassicaceae species, such as Thellungiella parvula and Aeyhionema arabicum. By providing a link between loss of ARC1 and self-compatibility, these results suggest that ARC1 functions in self-incompatibility signaling.
Exo70A1 was identified as an ARC1-interacting protein in a yeast two-hybrid screen (Samuel et al. 2009). Over-expression of Exo70A1 in the stigma epidermal cells of self-incompatible B. napus reportedly caused partial break-down of self-incompatibility. Furthermore, suppression of Exo70A1 expression in self-compatible B. napus and A. thaliana using RNAi and T-DNA insertions, respectively, reportedly resulted in inhibition of pollen adhesion, hydration, and germination (Samuel et al. 2009). Because Exo70A1 is a component of the exocyst complex, which generally functions in polarized secretion in yeast and animals ( Munson et al. 2006, Synek et al. 2006), the results suggested that factors required for compatible pollination are secreted to the outside of stigma epidermal cells through the exocyst complex. In self-incompatible plants, activation of SRK would cause activation of ARC1, which would ubiquitinate Exo70A1 and target it for degradation, thus preventing the secretion of factors required for proper hydration or germination of pollen grains.
Interestingly, very different results were obtained in an analysis of the role of the A. thaliana orthologs of MLPK, ARC1, and Exo70A1 using the transgenic A. thaliana SRK-SCR self-incompatible model (Kitashiba et al. 2011). A. thaliana contains a functional MLPK ortholog, AtAPK1b, which is located on chromosome 2 in a region that exhibits a high degree of synteny with the MLPK region of B. rapa. However, self-incompatibility was neither lost nor weakened in A. thaliana SRK-SCR plants that were homozygous for an AtAPK1b allele inactivated by T-DNA insertion (Kitashiba et al. 2011), indicating that a functional AtAPK1b is not required for self-incompatibility in the stigmas of A. thaliana SRK-SCR plants. In the case of ARC1, comparative analysis of the A. thaliana and A. lyrata genomes identified only fragmented sequences interspersed with other genes in the A. thaliana Col-0 and C24 accessions (Kitashiba et al. 2011), and the same fragmented organization of ARC1 sequences was also observed in all 96 A. thaliana accessions analyzed using PCR markers (Indriolo et al. 2012). In view of the fact that A. thaliana stigmas that express a functional SRK gene can exhibit an intense self-incompatibility response (Boggs et al. 2009a, Nasrallah 2002, 2004, Tsutimatsu et al. 2010), the results indicate that successful operation of the self-incompatibility signaling pathway in A. thaliana does not require a functional ARC1 protein. Nor does it require AtPUB17 (Rea et al. 2010), which is the member of the Plant U-Box family that exhibits the highest degree of sequence identity to B. napus ARC1 (Indriolo et al. 2012, Kitashiba et al. 2011).
Also contrary to the results obtained in B. napus (Samuel et al. 2009), overexpression of AtExo70A1 in stigma tissues of self-incompatible A. thaliana SRK-SCR plants did not weaken the self-incompatibility response (Kitashiba et al. 2011). Thus, it appears that the MLPK/ARC1/Exo70A1-based model of self-incompatibility signaling that was proposed for Brassica does not apply to the self-incompatibility response of A. thaliana SRK-SCR plants.
How might the discrepancies between the results obtained in Brassica and A. thaliana be reconciled? A possibility is that the experiments aimed at suppression of MLPK and ARC1 might not have been specific for the targeted genes and that A. thaliana genes related to the tested MLPK and ARC1 homologues might have assumed the role proposed for Brassica MLPK and ARC1 (Kitashiba et al. 2011). Another possibility is that self-incompatibility signaling is based, not on a single linear pathway, but on multiple signaling pathways (Tantikanjana et al. 2010), as reported for the plant defense response and signal transduction pathways in metazoans. These pathways might each contribute only partially to the overall self-incompatibility response, and different branches of the pathway might be utilized preferentially in different species of the Brassicaceae. Reexamination of the roles of the postulated signaling genes and their relatives in Brassica species and A. thaliana is required to distinguish between these possibilities.
Recently, mutagenesis of SRK-SCR transformants of the A. thaliana Col-0 accession revealed that a pathway involving small inhibitory RNAs controls self-incompatibility (Tantikanjana et al. 2009). Unlike SRK-SCR transformants of the C24, Cvi, Sha, Kas, and Hodja accessions, whose stigmas express an intense self-incompatibility response that persists throughout stigma development, Col-0[SRK-SCR] transformants express self-incompatibility in mature flower buds and very young flowers, but not in older stigmas. Because Col-0[SRK-SCR] transformants set seed, they were used for mutagenesis of the self-incompatibility trait using the standard approach of treating seed with a chemical mutagen (Tantikanjana et al. 2009). A screen for plants exhibiting a modified self-incompatibility response identified a mutation that enhanced the self-incompatibility phenotype of Col-0[SRK-SCR] transformants without affecting expression of the SRKb and SCRb transgenes (Tantikanjana et al. 2009). This mutation inactivated the RNA-dependent RNA polymerase RDR6, which functions in the production of trans-acing short interfering RNAs (ta-siRNAs). Interestingly, a mutation in ARGONAUTE7 ( AGO7), which functions downstream of RDR6 and is specifically responsible for the production of ta-siRNA targeting Auxin Response Factors (ARFs), also caused enhanced self-incompatibility in Col-0[SRK-SCR] transformants (Tantikanjana et al. 2009). Furthermore, overexpression experiments showed that ARF3, a member of the ARF family, acts non-cell-autonomously from its site of expression in cells below the stigma to enhance the self-incompatibility response and simultaneously down-regulate auxin responses (Tantikanjana and Nasrallah 2012). These results suggest the involvement of auxin in the regulation of self-incompatibility. Further analysis of mutants, either those that occur in natural populations (Isokawa et al. 2010) or those induced by mutagenic treatments (Strickler et al. 2013, Tantikanjana et al. 2009) will no doubt identify new components of the SRK-mediated signaling pathway and clarify the mechanism of the self-incompatibility response.
In the interspecific incompatibility observed between the pollen and pistil of different species, the inhibition of incompatible pollen is cytologically identical to that observed in the self-incompatibility response. For example, the inhibition of both conspecific “self” pollen and heterospecific pollen occurs during pollen tube growth within the style in the Solanaceae and by arrest of pollen germination and tube penetration into the stigma epidermal cell wall in the Brassicaceae. Interestingly, interspecific incompatibility is often unilateral (Lewis and Crowe 1958), i.e. the pistils of self-incompatible species generally inhibit pollen from self-compatible species, while the reciprocal pollinations are not inhibited. Genetic studies of unilateral incompatibility in tomato, a member of the Solanaceae, have revealed that this trait is controlled in the style by quantitative trait loci (QTL) (Bernacchi and Tanksley 1997, Li and Chetelat 2010), one of which corresponds to the S-RNase gene, which is the stylar determinant of self-incompatibility in the Solanaceae (Bernacchi and Tanksley 1997). Furthermore, two genes encoding pollen factors involved in interspecific incompatibility were identified: one gene was linked to the S locus on chromosome 1, while the other was located on chromosome 6 (Chetelat and De Verna 1991) and found to be Cullin 1, which is highly similar to a pollen factor that functions in the self-incompatibility response in Petunia (Li and Chatelat 2010). These results identify substantial overlaps in the pathways that underlie pollen inhibition in self-incompatibility and interspecific incompatibility in the Solanaceae.
In the Brassicaceae, interspecific incompatibility has been observed in interspecific pollinations between B. rapa and B. oleracea. In addition, one case of unusual unilateral intraspecific incompatibility was reported in B. rapa (Takada et al. 2013), which will not be discussed further. In all cases of interspecific unilateral incompatibility that have been examined, only cytological and genetic analyses have been performed, and the molecular basis of pollen inhibition is not known. Nevertheless, some studies have even invoked the involvement of the S locus in unilateral interspecific incompatibility (Hiscock and Dickinson 1993).
Genetic analysis of Brassica interspecific incompatibility has provided some information on the stigma’s ability to discriminate against heterospecific pollen, but as yet nothing is known about pollen factors that might be involved in this process. Udagawa et al. (2010) identified two strains of B. rapa that differed in the response of their stigmas towards heterospecific B. oleracea pollen: one strain exhibited a strong interspecific incompatibility response and its stigmas were highly inhibitory to B. oleracea pollen, while the other strain did not exhibit interspecific incompatibility and its stigmas allowed profuse pollen tube growth when pollinated with B. oleracea pollen. The interspecific incompatibility trait in stigmas was found to be dominant and controlled by quantitative trait loci (QTL). Five QTL explained over 75% of the phenotypic variance between the two strains, with one QTL on linkage group 2 explaining 32% of this variance. Contrary to the expectation that interspecific incompatibility might be based on the activity of ARC1 as proposed for self-incompatibility in Brassica, only a peak with non-significant LOD score was observed at the ARC1 locus. Moreover, in another B. rapa population derived from a cross between the self-compatible ‘Yellow Sarson’ strain and a self-incompatible strain, the strength of interspecific incompatibility was independent of genotypic composition at the M locus and its MLPK gene (Udagawa et al. 2010). Thus, no evidence has been found so far for the sharing of components between the self-incompatibility and interspecific signaling pathways in Brassica. A firm conclusion on this issue must await the molecular cloning of genes underlying the interspecific incompatibility QTL as well as of genes that encode components of the self-incompatibility pathway.
Despite the lack of supporting data, the parallels between pollen inhibition in self-incompatibility and interspecific incompatibility observed among species of the Brassicaceae are suggestive of the existence of a stigma-based response pathway that is shared, at least in part, by the two incompatibility systems. Clearly however, the recognition of “self” pollen and of heterospecific pollen must be mediated by distinct factors in stigma and pollen. Although the identity of these factors is not known, it may be assumed that, unlike SCR, which induces the self-incompatibility response, any factor in heterospecific pollen that might trigger an incompatibility response in the stigma must exhibit limited intraspecific polymorphism but extensive polymorphism between species and genera. This assumption is based on the fact that interspecific incompatibility analyzed in B. rapa is typically characterized by a uniform response to heterospecific pollen, irrespective of its origin. Thus, in the Udawaga et al. (2010) study, the pollen of all B. oleracea strains tested was inhibited on the stigmas of the B. rapa strain that expressed interspecific incompatibility but not on the stigmas of the strain that did not exhibit interspecific incompatibility. Moreover, B. rapa strains that express the interspecific incompatibility trait uniformly inhibited pollen derived from plants belonging to several species and genera of the Brassicaceae, such as B. nigra, R. sativus, Eruca sativa (our unpublished data). And all B. rapa strains analyzed in these studies were compatible with conspecific pollen derived from plants that express different S haplotypes (Udagawa et al. 2010).
Here we present a highly speculative model of a universal pollen-inhibition pathway that is shared by self-incompatibility and interspecific incompatibility. The model makes the following assumptions: (1) interspecific incompatibility involves recognition of a species-specific pollen factor by a stigma factor; (2) the stigma factor responsible for recognition of heterospecific pollen is a receptor protein kinase which is located in the plasma membrane of stigma epidermal cells; (3) the interaction between the stigma receptor and the species-specific pollen factor activates the same “universal” signaling pathway that causes inhibition of pollen in the self-incompatibility response, and (4) the factor found in conspecific pollen is a negative regulator of receptor activity. As shown in Fig. 2, in unpollinated stigmas, the receptor would be active and the universal pollen-inhibition pathway would be activated, i.e. it is in the ‘On’ state. This pathway will remain ‘On’ in the presence of heterospecific pollen because the receptor cannot recognize the pollen factor from other species. However, in pollinations with conspecific “nonself” pollen (i.e. pollen derived from plants of the same species that express S haplotypes that are different from those expressed in the stigma), the receptor kinase recognizes the conspecific pollen factor, whereby it is inactivated, causing the pollen-inhibition pathway to be turned to the ‘Off’ state and allowing pollen tube development.
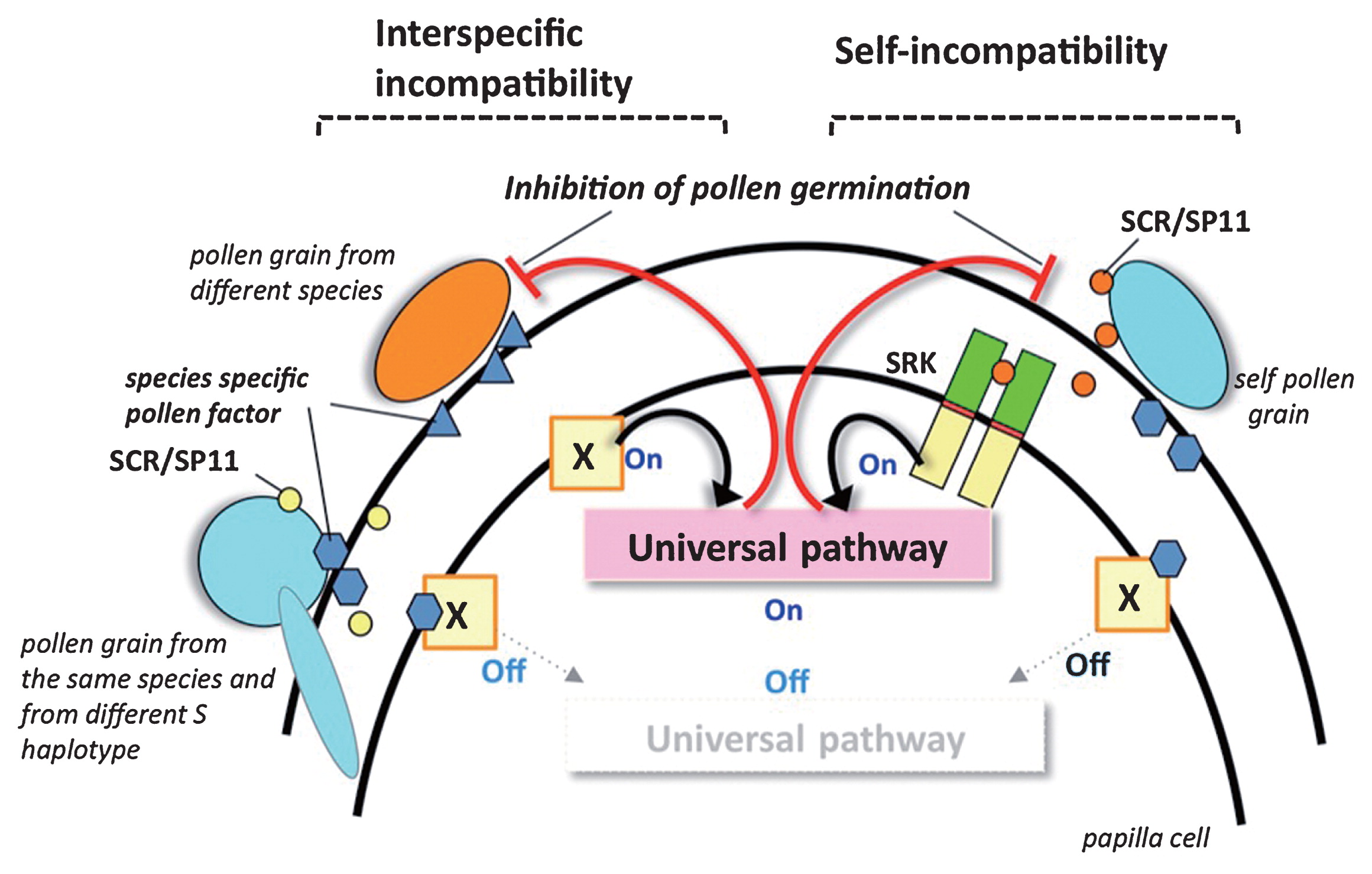
Model of signaling thorough a hypothesized universal pathway in Brassica self-incompatibility and interspecific incompatibility. X is a hypothesized receptor kinase.
Notably, this hypothetical scheme differs substantially from the accepted mode of SRK activation in the self-incompatibility response. Whereas SRK, which is inactive until bound by its cognate SCR, the receptor kinase proposed for interspecific incompatibility is postulated to be constitutively active and to be inactivated by interaction with a species-specific pollen factor. This model of negative regulation of receptor activity overcomes some of the issues associated with a model based on receptor activation by a species-specific pollen factor, which would require the existence in the stigma of a large number of receptors, each of which would recognize, and become activated by, a pollen factor specific to a particular species. In any case, the proposed mode of receptor regulation resembles the regulation of the ethylene receptor ETR1, which is constitutively active in the absence of ethylene and switched off in the presence of ethylene (Yoo et al. 2009).
What might the identity of the postulated receptor be? The genomes of Brassicaceae and other plant species contain large numbers of genes encoding receptor kinases that are similar to SRK in sequence and structure (Shiu et al. 2004). For example, 38 such genes are found in the A. thaliana genome (Xing et al. 2013). It is possible that one of these SRK-like receptor kinases might function as a stigma receptor in interspecific incompatibility. It is hoped that continued molecular genetic analysis of Brassica and Arabidopsis species will elucidate the mechanisms that underlie pollen inhibition in interspecific incompatibility and self-incompatibility and clarify any overlaps that might exist between mechanisms of pollen inhibition in the two systems.