2015 年 65 巻 3 号 p. 285-289
2015 年 65 巻 3 号 p. 285-289
More accurate, rapid, and easy phenotyping tools are required to match the recent advances in high-throughput genotyping for accelerating breeding and genetic analysis. The conventional data recording in field notebooks and then inputting data to computers for further analysis is inefficient, time-consuming, laborious, and prone to human error. Here, we report WIPPER (for Wireless Plant Phenotyper), a new phenotyping platform that combines field phenotyping and data recording with the aid of Bluetooth communication, thus saving time and labor not only for field data recoding but also for inputting data to computers. Additionally, it eliminates the risk of human error associated with phenotyping and inputting data. We applied WIPPER to 100 individuals of a rice recombinant inbred line (RIL) for measuring leaf width and relative chlorophyll content (SPAD value), and were able to record an accurate data in a significantly reduced time compared with the conventional method of data collection. We are currently using WIPPER for routine management of rice germplasm including recording and documenting information on phenotypic data, seeds, and DNA for their accelerated utilization in crop breeding.
For many crop species, considerable wild and landrace germplasms have been collected globally to expedite utilization of the available diversity for crop improvement as well as for conservation purposes (Esquinas-Alcázar 2005). Additionally, new breeding resources such as mutant lines and various mapping populations are being developed to enhance the utilization of genetic diversity in breeding and genetic analysis (Cavanagh et al. 2008). Large-scale analysis of crop germplasm has become possible following the recent advances in next generation sequencing (NGS) technologies that revolutionized whole genome sequencing (WGS) of plants.
For example, the recently developed WGS-based methods of MutMap and its derivatives allow mapping of genes or QTLs of interest in a relatively short time (Abe et al. 2012, Fekih et al. 2013, Takagi et al. 2013). Once a mutant of interest is identified, MutMap enables rapid isolation of the candidate gene even when the phenotypic changes are only subtle. Additionally, novel NGS-based genotyping methods such as restriction-site associated DNA (RAD) sequencing (Yada et al. 2011) and genotyping-by-sequencing (GBS) (Robert et al. 2011) have enabled high-throughput genotyping in crops including those with large genome sizes, for which the application of WGS remains difficult and expensive. For instance, GBS has been recently applied to 5,000 recombinant inbred lines (RILs) for large-scale association mapping in maize (Kump et al. 2011), highlighting the need for large-scale phenotyping protocols for accurate association of genotype-phenotype data.
While genotyping protocols have advanced as a result of the technological progress achieved in WGS, establishing high-throughput phenotyping methods for quantifying traits of interest has remained challenging. Conventionally, researcher record phenotype data into field notebooks, and the recorded data are inputted into PC by hand typing for further analysis. This procedure is both time-consuming and labor-intensive. Particularly, recording data under difficult field conditions such as in rice paddy field is not a straightforward task and is often associated with human errors. When dealing with a large data set, there is an additional risk of introducing error due to mistyping. Consequently, although WGS and genotyping have become routine procedures, genetic analysis and breeding are still constrained by lack of accurate and high-throughput phenotyping protocols.
In this study, we developed an accurate phenotyping method called WIPPER that is suitable for large-scale applications in the field to aid research in various crops including accelerating genetic analysis. WIPPER combines the use of personal devices assistant and Bluetooth communication to reduce the time required for and increase the accuracy or efficiency of data recording. We demonstrated the applicability of WIPPER for large-scale field phenotyping by comparing its performance with the conventional method of data recording into field notebooks for phenotyping of leaf width and SPAD value of rice mapping population.
The quick response (QR) type barcode label for identifying individual plants or lines was generated by the Barcode Sakusei Kanri Kun Pro software (NCE, Japan). The QR barcodes were printed on Kokuyo KB-A2190 waterproof sheets (Kokuyo, Japan) that are durable at least for three years under field conditions. The code, bearing the name of each individual or line, was represented by 10 numeric characters that include information on line number, year of harvest, and crossing combination.
Data measurement and recordingData on leaf width was recorded electronically using a digital caliper (Mitsutoyo Digimatic Caliper CD-15CX, Mitsutoyo, Japan) that was connected with Digitec Bluetooth adapter DKA-101 (digi-tek, Japan). The data measured by a digital caliper were transported via Bluetooth communication to the barcode reader PRAViON PM-250TA, which was installed with the software “Rice Phenotyping System for Mobile Device” (FCR bio-company, Japan) enabling it to receive data from digital measurement with Bluetooth communication, such as a digital caliper in this study. All the data recorded in barcode reader were transferred via a USB connector to a PC on which the software “Rice Phenotyping System for PC” was also installed.
For measuring SPAD value, which is proportional to the relative leaf chlorophyll contents, SPAD-502bt (KONICA MINOLTA INC, Japan) was used. SPAD-502bt was developed from SPAD-502 with the added capacity for Bluetooth communication custom made for this research. SPAD values representing the mean of three readings were immediately transferred into a smart phone (HTL22, HTC, Taiwan) that runs on the android operation system and installed with a dedicated software Spad1.2 (KONICA MINOLTA INC) for immediately receiving data from SPAD502bt via Bluetooth communication. The SPAD value data are finally transferred into Cloud storage server designated to each user. BOX server (https://www.box.com/) was used for data storage in this study.
WIPPER is a new field phenotyping system that is aided by wireless communications for data recording and storage as described in Fig. 1. In WIPPER, to identify individual lines or accessions, their barcodes are first scanned with a suitable personal digital assistant (PDA) including barcode reader, cellular phone, tablet PC and so on. Then, the phenotype data are collected using electronic measuring devices, e.g. electronic scale, electronic balance, and digital angle meter equipped with Bluetooth wireless communication system, which enables data transfer from electric measuring devices to PDA. Finally, the collected data in PDA are exported to a storage environment (PC or storage server) by USB connecter or Wi-Fi. By eliminating the need for data recording into notebook and manual data transfer to PC, WIPPER significantly minimizes or eliminates human error and improves the accuracy of data collected.
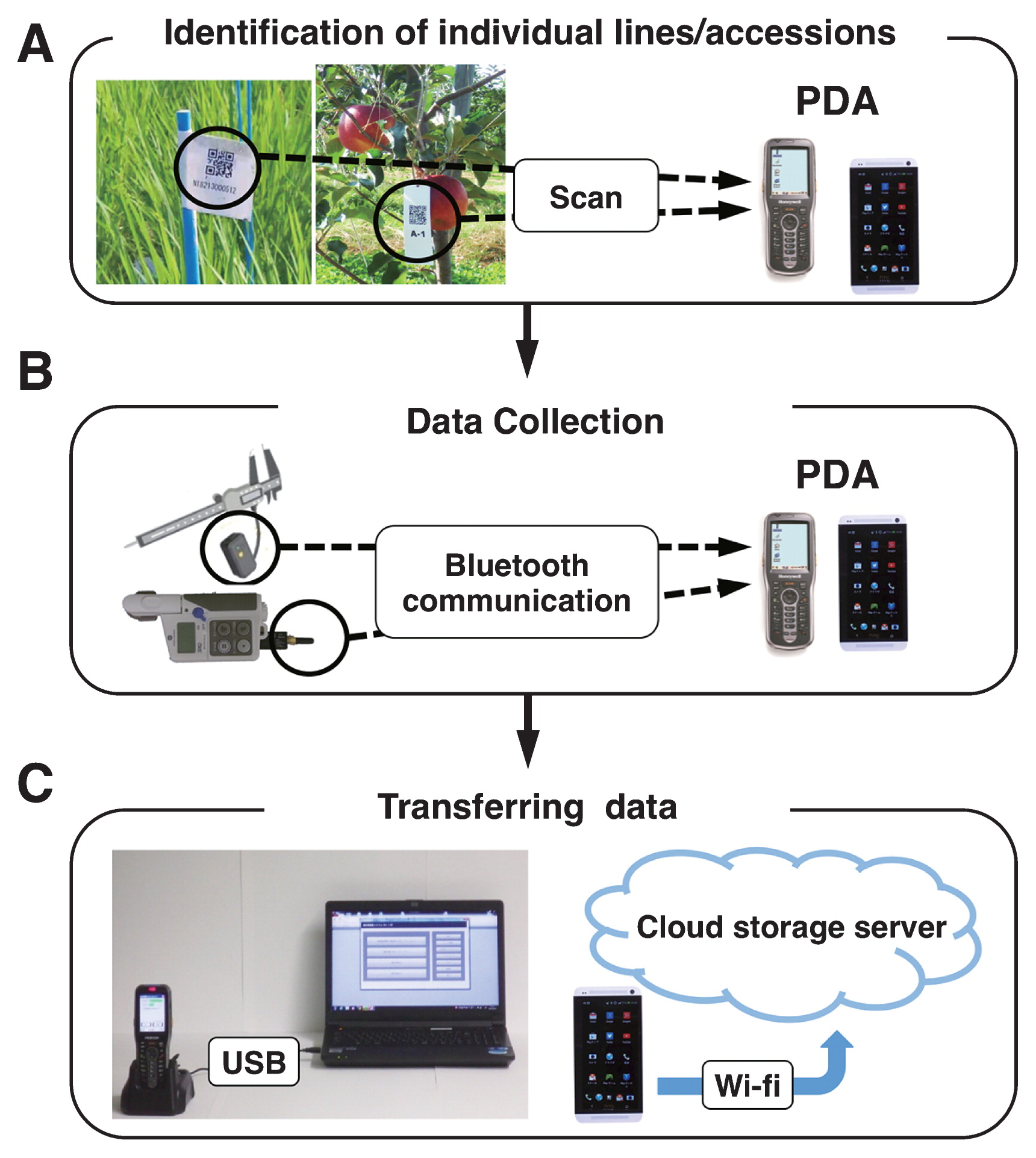
Flowchart of WIPPER. (A) A personal devise assistant (PDA) identifies the line/accession to be phenotyped upon scanning its barcode. (B) Data are collected by PDA via Bluetooth wireless communication. Black circles indicate the Bluetooth connector enabling each electric or digital phenotyping device to transfer data to the PDA by Bluetooth communication. (C) The data in PDA are exported to a storage environment via a USB connecter or Wi-Fi.
To demonstrate the utility of WIPPER, we applied it for measuring leaf width and compared its efficiency with the conventional method of data collection by handwriting into field notebooks as illustrated in Fig. 2. First, individual lines or plants were identified by scanning their barcode label using the barcode reader (Fig. 2A). Then, data on leaf width were collected by a digital caliper (Mitsutoyo Digimatic Caliper CD-15CX) equipped with Bluetooth connector (Fig. 2B). The data were immediately transferred into barcode reader by pressing the button for Bluetooth wireless communication. The data were finally exported from barcode reader to PC via USB connector (Fig. 1C). In the conventional method, each line number and leaf widths of the corresponding individuals were recorded by handwriting to notebook, which was then inputted into PC by hand typing. We also applied the conventional method either with one person or two people working together. When two people were involved, one of them measured leaf width while the other person wrote down the data into a field notebook.
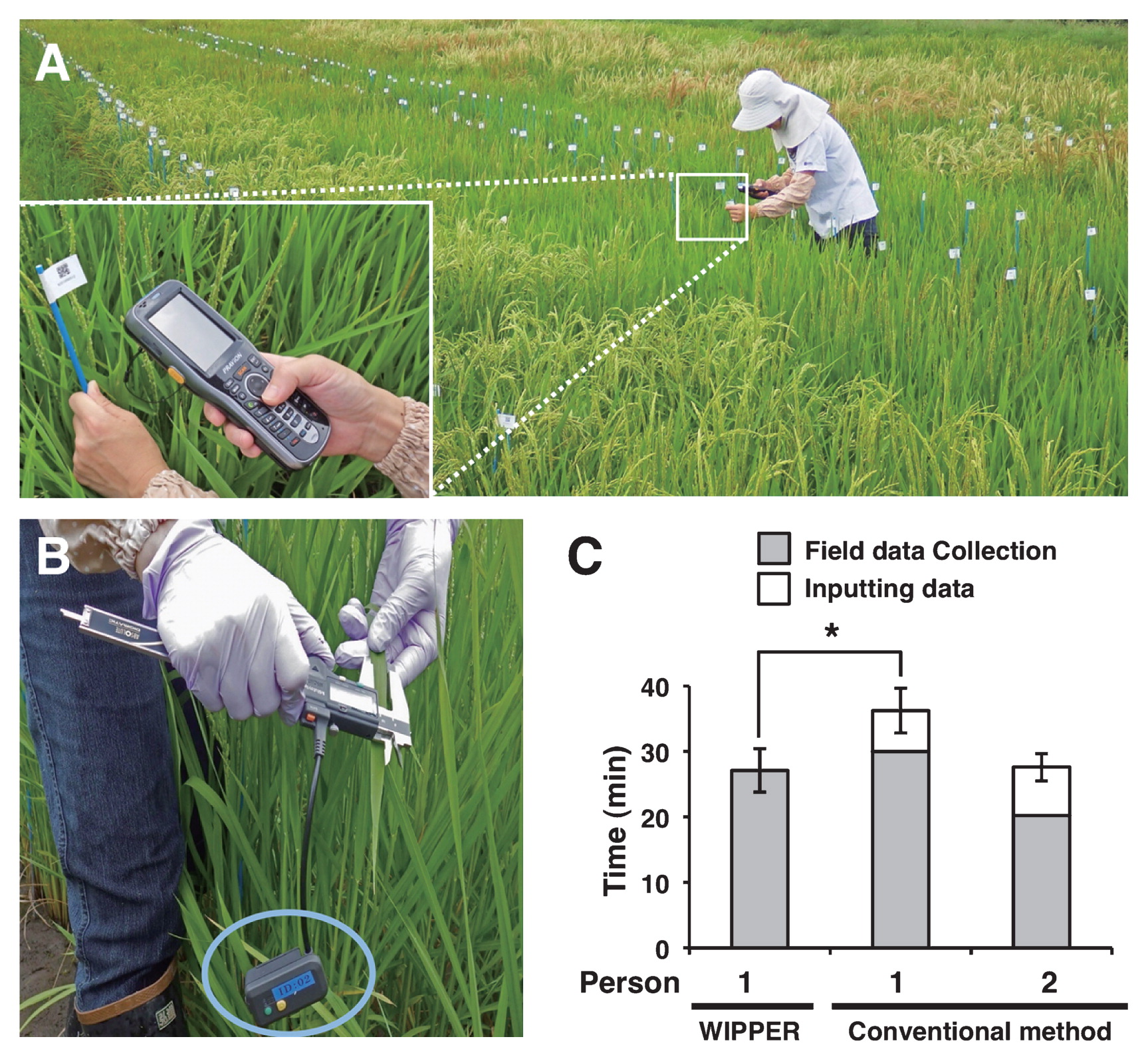
Measuring leaf width in rice recombinant inbred lines by WIPPER. (A) Identification of each line/accession by scanning with a Barcode reader. (B) A digital caliper (Mitsutoyo Digimatic Caliper CD-15CX) with Bluetooth connector is used for measuring leaf width. Blue circle indicates the Bluetooth connector. (C) Comparison of the time required (in minutes) for measuring leaf width in 100 (5 individuals per each of the 20 lines) by WIPPER and the conventional method involving manual data collection and inputting by one person or two people. Asterisks indicate significant difference (Student’s t-test, * P < 0.05; normality of variables and equality of variances were validated by Kolmogorov-Smirnov test and F-test, respectively).
A total of 100 individuals representing 20 RILs (five plants per line) were measured by both methods, and the total time required for data collection and inputting was compared. The total time required by the conventional method involving one person, two people, and one person using by WIPPER was 36.3 min, 27.6 min, and 27.1 min, respectively. WIPPER successfully reduced the working time by 25.3% compared with the time required to accomplish the same task by the conventional method involving one person (Fig. 2C). Although WIPPER required more time for measuring leaf width in the filed than the conventional method involving two people, the total working time needed was still shorter due to the fact that WIPPER did not require additional time for data inputting into PC, as this was done simultaneously via a USB connector. In the conventional method, it took more than 6 minutes for inputting the data on 100 individuals into a PC not mentioning the potential human error associated with such a task. Overall, WIPPER had the advantage for reducing the working time and increasing the data accuracy, and this will have significant implications when considering large-scale application of WIPPER in the field.
WIPPER applied for measuring leaf relative chlorophyll contents using a SPAD meterBecause the amount and time of availability of essential plant nutrients determine crop growth and yield, SPAD value is usually used as an important indicator of the relative chlorophyll content of leaves for timing fertilizer application in crop fields including rice (Fred et al. 1991). To this end, SPAD value has to be continuously monitored during the growing period. For effective monitoring of SPAD values, we also applied the WIPPER system as described in Fig. 3. In this investigation, mobile devices such as smart phone and tablet PC were used as substitutes for Barcode reader (Fig. 3A). Accordingly, the mobile devices were first used to scan the bar code labels similar to the way it was done for leaf width measurement. Next, SPAD values measured with the modified SPAD-502 meter equipped with Bluetooth connector were automatically transferred into the mobile device by Bluetooth wireless communication (Fig. 3B). Finally, the SPAD values recorded in mobile device were transferred automatically into cloud storage server, which was designated to each user in Wi-Fi environment (Fig. 3C).
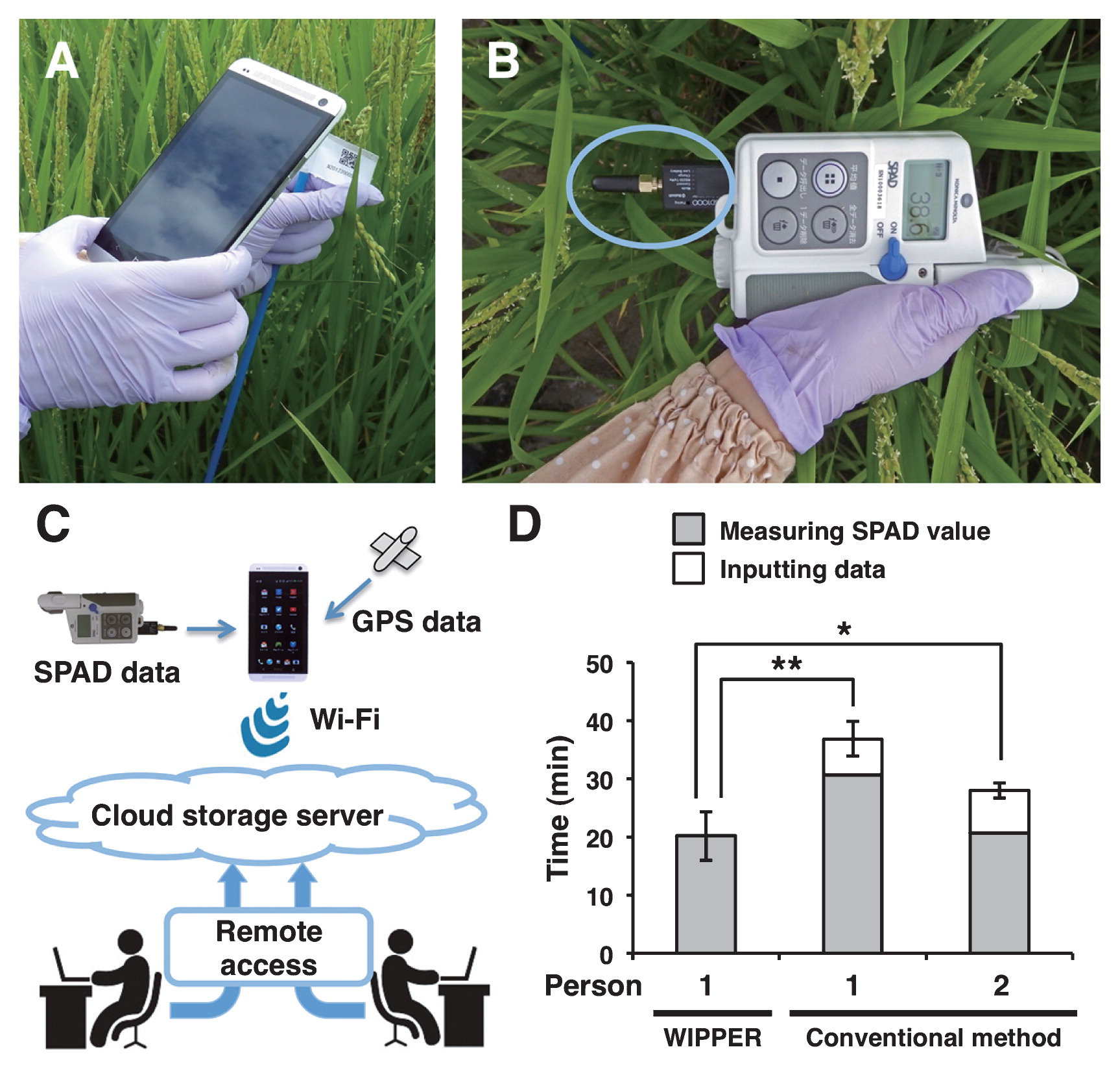
Measuring SPAD value in rice recombinant inbred lines by WIPPER. (A) Identifying each line/accession by scanning its corresponding barcode using a smart phone. (B) An SPAD502 meter with Bluetooth connector is used for measuring SPAD values. Blue circle indicates Bluetooth connector. (C) Scheme of the cloud storage server applied in this study. (D) Comparison of the time taken (in minutes) for measuring SPAD values of 100 individual plants (5 individuals from each of 20 lines). Asterisks indicate significant difference (Student’s t-test, * P < 0.05, ** P < 0.01; normality of variables and equality of variances were validated by Kolmogorov-Smirnov test and F-test, respectively).
Again, we compared the working time requirement of WIPPER with the conventional methods involving either one person or two people for collecting and inputting SPAD values. The working time of the conventional methods by one person and two people was 36.9 min and 28.0, respectively, while WIPPER required 20.2 min to accomplish the same task. This corresponded to 45.3% and 27.9% less total working time required by WIPPER compared with the conventional method by one person and two people, respectively (Fig. 3D). Notably, the time required for measuring SPAD value was significantly lower compared with the time it took for measuring leaf width. This was primarily because the application software used in SPAD-502bt allows data transfer to smart phone without the need to press a button, while the one used with the digital caliper for measuring leaf width required pressing a button after each measurement for transferring data. Additionally, as the SPAD values were stored in a cloud storage server, the risk of human error in data recording was completely eliminated. The use of cloud storage server also means unlimited number of users could access the data from remote location in real time. Overall, WIPPER reduced the working time while increasing the data accuracy.
Recent advance in genotyping methods such as Illumina GoldenGate assay that allow accurate and high-throughput genotyping are revolutionizing crop breeding as well as genetic analysis (Akhunov et al. 2009). For instance, using a large number of genotyping data from GoldenGate assay (Buckler et al. 2009) carried out a genetic analysis named nested association mapping (NAM) to dissect the genetic architecture of flowering time in maize. While genotyping methods have improved, phenotyping has largely remained conventional, and currently represents the main bottleneck in achieving the necessary accuracy and speed in genetic analyses. The WIPPER system described in this study has proved effective in reducing the time and labor required not only for field phenotyping but also for inputting data into PC, while significantly reducing or eliminating the human error commonly associated with conventional phenotyping, thereby ensuring accuracy of phenotype data. We therefore believe that WIPPER is applicable for any analysis involving the collection of large-scale phenotype data.
Although we only presented two examples as proof-of-principle applications, the concept of WIPPER could be adapted to any digital measurement that researchers want to use. For example, an electric balance with Bluetooth connection can be used for recording yield or data on weight. Similarly, a digital angle meter equipped with Bluetooth can be adapted for measuring shoot or tiller angle in many crops. We also believe that the application of WIPPER can improve the efficiency of many of the recently developed high-throughput phenotyping tools that target leaf shape (Bylesjö et al. 2008), panicle structure (Faroq et al. 2013) and root structure (Pace et al. 2014), which involve data measurement after sample collection. If these tools can be applied in the field, the WIPPER platform reported here could be adapted to improve the efficiency of data recording. On the other hand, although the recently reported phenotypic platform named FieldBook (Yamasaki and Garcia 2012) is similar to WIPPER in that it also uses the barcode system, it lacks the flexibility of WIPPER, which makes use of Bluetooth communication for data transfer thereby allowing high-throughput phenotyping. In general, WIPPER is a versatile platform with a wider applicability for phenotyping with digital measurements, providing flexibility and reliability to various researchers working in the field.
For measuring SPAD value, we used a smart phone that enabled us to also record the exact location of the experimental field by importing Global Positioning System (GPS) information. Although resolution of the location depends on the performance of GPS in the smart phone, the one in the smart phone used for this study had a high-resolution with an accuracy of about one meter. The possibility of integrating GPS information to the WIPPER system implies that the scanning step for identifying individual plants to be phenotyped could be skipped if the GPS is of high enough resolution to discriminate individuals and if the information of the location was inputted in advance. This has practical applicability for researchers who would like to phenotype trees by this system, as phenotyping can be done using only a device with Bluetooth connector. The data for each tree could be retrieved using the corresponding GPS information. If such a system is established, it will broaden the applicability of WIPPER. Additionally, the system of transporting data to cloud storage server, which we used with the SPAD values recorded, allows researchers to centralize data analysis. This centralized data management makes it possible for one researcher to analyze the data simultaneously collected by different investigators in multiple locations.
Currently, we identify our rice germplasm including seeds and DNA samples of cultivated rice, wild relatives, and mutant populations using the same accession numbers given to each line for phenotyping. This involves packing seeds in barcoded envelopes or bags, as well as storing DNA samples in barcoded individual tubes (Fig. 4A, 4B). Our system ensures that researchers can easily access seeds or DNA samples of the individuals they want to further investigate based on phenotypes of interest. In future, we are planning to add genotyping data including whole genome sequence to the system (Fig. 4C). We believe that such systematic integration of relevant data will speed up genetic analysis and accelerate crop breeding, as well as improve cultivation techniques.
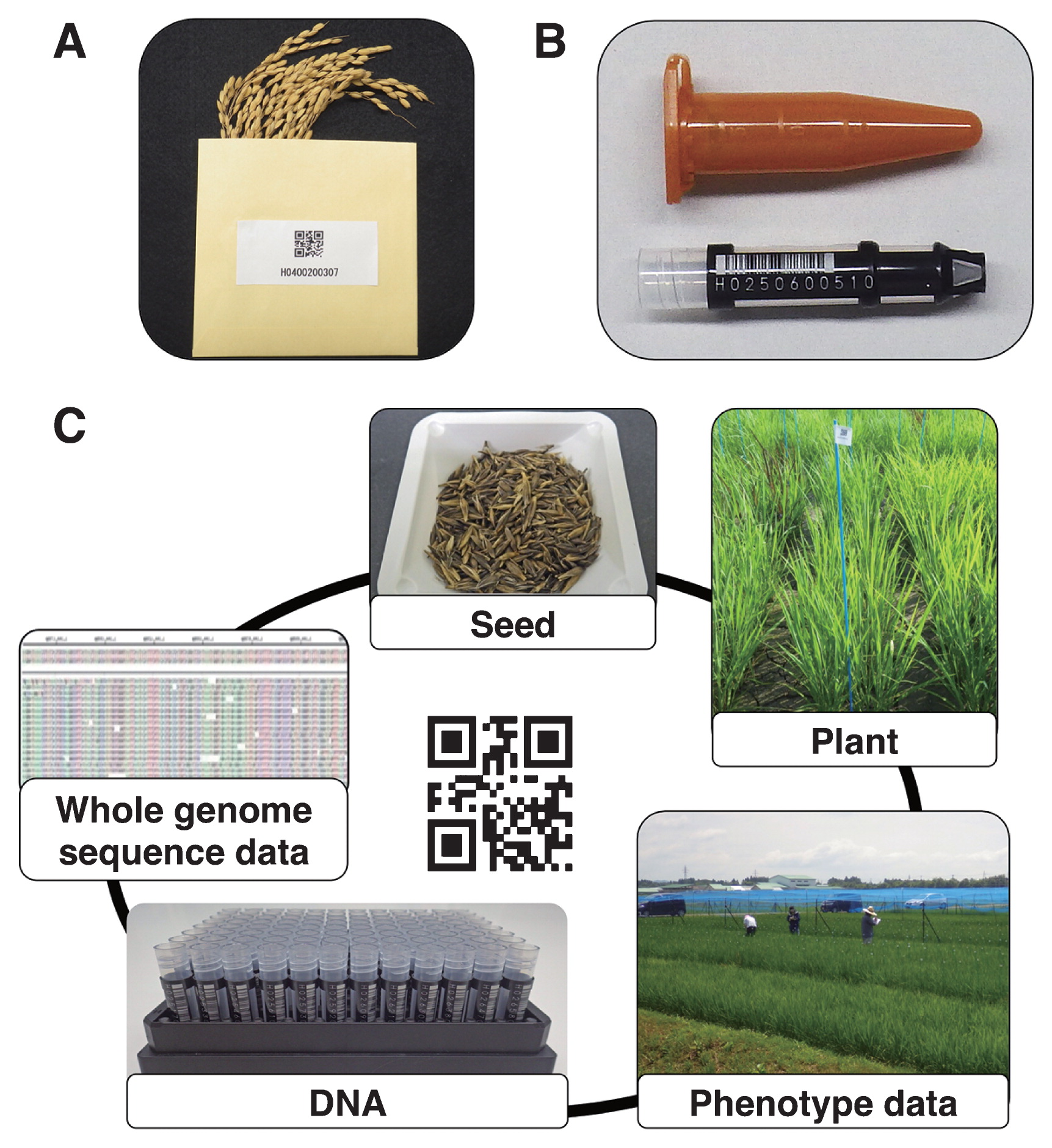
The application of WIPPER for an integrated germplasm and data management towards rapid genetic analysis. (A) Rice seeds stored in envelopes. Each envelope is labeled with a barcode containing all the necessary information about the line/accession. (B) DNA samples stored in barcoded tubes. (C) The concept of an integrated data management. For each line/accession, the same barcode or number is used to identify its phenotypic data, seeds, DNA sample, as well as genotyping data including whole genome sequences.
We acknowledge the help we received from Hiroshi Yamamoto (FCR bio Co. Ltd, Japan) and Shinji Takashige (Imager Co. Ltd, Japan), and Tomomi Setoguchi (KONICA MINOLTA INC, Japan) and Takuya Matsumoto (KONICA MINOLTA INC, Japan) in developing the system to measure leaf width and SPAD value by WIPPER, respectively. This work was supported by JSPS KAKENHI Grant Number 26712002 to AA, as well as by Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry, Japan, MEXT KAKENHI (Scientific Research on Innovative Areas 23113009) grant to RT.