2017 Volume 67 Issue 2 Pages 110-122
2017 Volume 67 Issue 2 Pages 110-122
Fruit brown rot caused by Monilinia spp. is the most important fungal disease of stone fruits worldwide. Several phenotyping protocols to accurately characterize and evaluate brown rot infection have been proposed; however, the outcomes from those studies have not led to consistent advances in resistance breeding programs. Breeding for disease resistance is one of the most challenging objectives for crop improvement because disease expression is tetrahedral: it is simultaneously influenced by agent, host, environment, and human management. The present study presents a strategy based on Bayesian inference to analyze a peach breeding progeny for resistance to brown rot, evaluated using a polytomous ordinal scale. A pedigree containing two sources of resistance, one from peach and the other from almond, several commercial cultivars, and two segregating populations were analyzed to estimate the narrow-sense heritability (h2) and breeding values (EBVs) for brown rot resistance in progenies. Results show promise for genetic improvement of disease resistance and other traits characterized by strong environmental interactions.
Brown rot (BR) is the general term given to the symptoms of infection and sporulation on stone fruit, primarily by Monilinia fructicola (G.Winter) Honey, and to a lesser extent by Monilinia laxa (Aderh. & Ruhland) and Monilinia fructigena Honey. BR is the most important disease of peach worldwide (Gradziel 2003, Ogawa and English 1991) and the frequent application of fungicides required to control it remains a major constraint to sustainable production. Consequently, genetic resistance to BR is highly desirable in new cultivars.
Various resistance and tolerance germplasm sources are known (Byrne et al. 2012), and several efforts have examined the way in which M. fructicola infects fruits (Lee and Bostock 2007, Lee et al. 2010). Although fruit traits associated with resistance (Gradziel 2003) and part of their genetic components (Martínez-García et al. 2013) have been characterized, successful breeding of resistant commercial cultivars remains elusive, due in part to the strong environmental influence, the absence of a focused research effort including the development of an efficient evaluation criteria. Characteristics of the fruit epidermis, such as thickness of the cuticle and epicuticular wax layer, and concentrations of pectin, phenolic, chlorophyll, and other biochemical compounds associated with tissue maturation (Gradziel and Wang 1993, Lee and Bostock 2007), are associated with the degree of resistance within a given phenotype when the fruits are not wounded. Epidermal barriers can be lost following mechanical or insect damage, leaving only mesocarp resistance factors such as high phenolic concentration or flesh textural integrity, which may be commercially undesirable (Gradziel 2003).
Procedures for characterizing epidermal characteristics are labor intensive, expensive, and inconsistent (Byrne et al. 2012). In wounded fruit, where the epidermis has been breached, Martínez-García et al. (2013) identified three putative quantitative trait loci (QTLs) and two candidate genes associated with the BR resistance. However, because the phenotypic data and residuals distribution from non-wounded fruit exhibited serious departures from a normal distribution due to serious departures from homoscedasticity (i.e. heterogeneity of variance), the data was not used for QTL analysis, which assumes homoscedasticity. In this case, data transformation would not help to alleviate the issues while not altering intuitiveness of the phenotypic scale during conclusion drawing. Information generated through artificial inoculation on non-wounded fruit is considered more reliable and representative of BR field resistance (Byrne et al. 2012).
In peaches bred for canning, normally undesirable fruit epidermis-based resistance such as high phenolic concentration and cellular integrity can be targeted, since the epidermis is removed during processing. Towards this goal, sources of epidermis-based resistance from peach, including the resistant cultivar ‘Bolinha’ (Feliciano et al. 1987), as well as from almond and related species have been incorporated into advanced breeding selections, through genetic introgression (Gradziel 2002, Gradziel et al. 2003). While BR resistance has been improved, the derived germplasm often shows reduced quality for important commercial traits as a result of linkage drag (Gradziel 2003, Gradziel and Wang 1993).
Concurrent selection for epidermis-based resistance and good fruit quality is consequently highly desirable, despite the genetic complexity of these traits and their often exotic origins. Towards this goal, a strategy based on Bayesian inference is presented as an application of the individual model (also known as “the animal model” in animal breeding literature), referred to here as the Bayesian ordinal animal model (BOAM). A pedigree-based narrow-sense heritability (h2) was estimated and the estimated breeding values (EBVs) were obtained by fitting ordinal logistic regression by means of the Gibbs sampler integrated in JAGS (Plummer 2003). To evaluate the robustness of BOAM, the evaluation of the goodness-of-fit was performed for several models, using distinct priors for the probability distribution of the square root of the additive genetic variance
The pedigree analyzed is composed of 221 accessions, including processing peach cultivars and breeding selections, the almond cultivar ‘Nonpareil’, and two segregating mapping populations (PopBR-1 and PopBR-2) resulting from the crosses ‘Dr. Davis’ × ‘F8,1-42’ and ‘Loadel’ × ‘96,4-55’, respectively (Fig. 1). The first cross, involved a resistant modern cultivar and a breeding line possessing BR resistance from almond, respectively. The second cross, involved a resistant old cultivar and a breeding line possessing BR resistance from the Brazilian cultivar ‘Bolinha’, respectively.
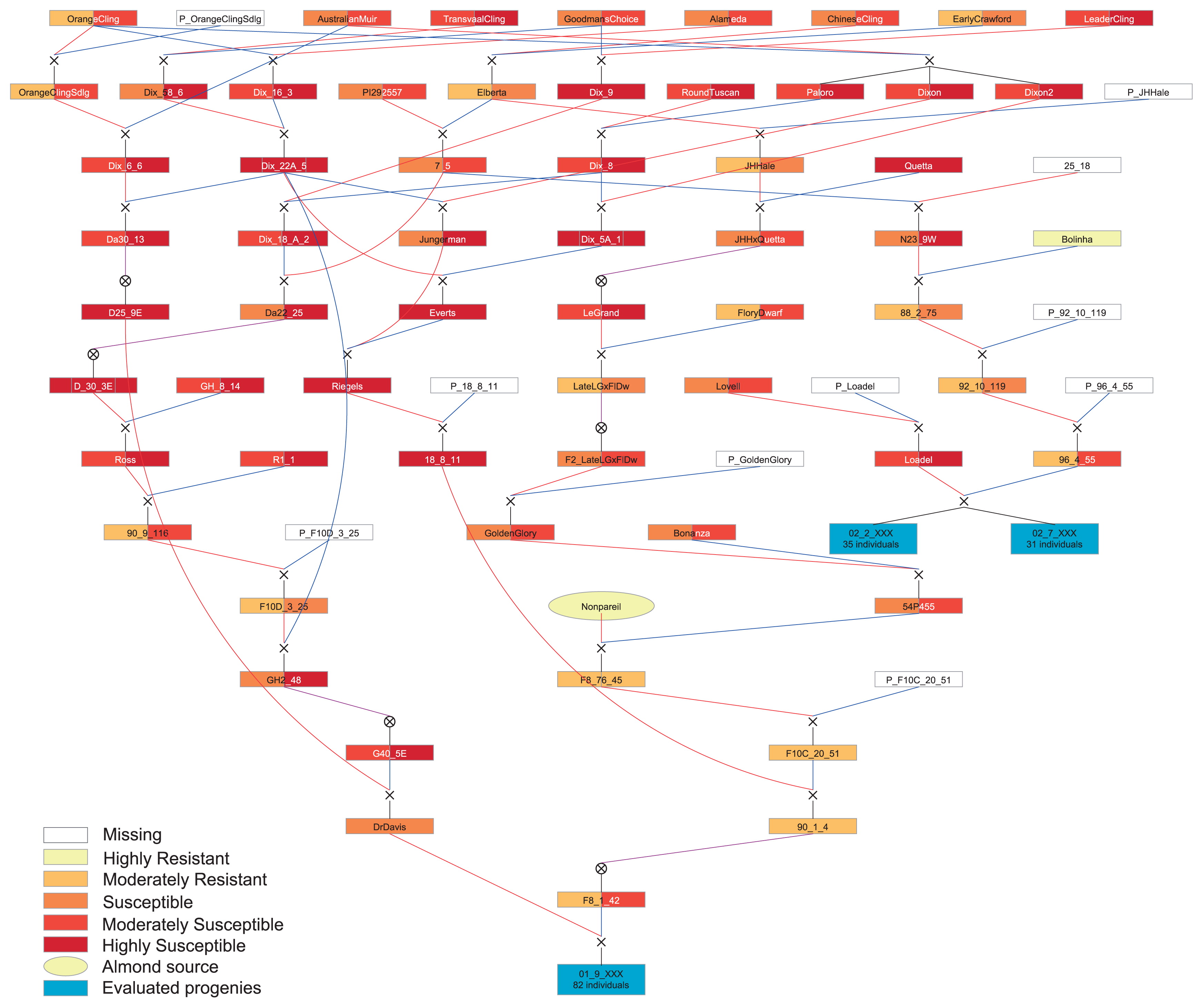
Pedigree of the accessions studied. Each box represents the degree of BR resistance in non-wounded (left) and wounded (right) fruits. In light yellow are the almond cultivar ‘Nonpareil’ (oval), the source of BR resistance from almond, and ‘Bolinha’ (rectangle), the source of BR resistance from peach, and in blue are the progenies PopBR-1 (01 09 XXX) and PopBR-2 (02 02 XXX and 02 07 XXX). Empty boxes mean no information is available for that accession, usually unknown male parentage. The pedigree figure was generated using PediMap 1.2 (Voorrips et al. 2012) and colored through Adobe Illustrator CS5.5.
A severity index was determined for the progenies of the breeding crosses, as described by Martínez-García et al. (2013). In summary, twenty fruits at the tree-ripe maturity stage from a single individual were collected during each season, which extended over a 6 to 7 week period of July and August, during the 2007–2009 seasons. Fruit maturity differed slightly from season to season. The fruits were stored at 4 °C, for four days and subsequently warmed to room temperature for 24 h prior to inoculation. Fruit outer surfaces were sterilized for 30 seconds through immersion in 10% bleach (0.6% NaOCl), rinsed in deionized water, and dried. Fruits were then placed in humidified plastic containers (30.5 cm × 22.9 cm ×10.2 cm, Model 295C; Pioneer Plastics, Dixon, KY) with fruit tray liners (M-24B; FDS Manufacturing Co., Inc., Pomona, CA). Fruits were inoculated with a 10 μL droplet containing conidia of M. fructicola (isolate MUK-1) at a concentration of 2.5 × 104 spores per mL from 7 to 10-day-old cultures maintained on V-8 juice agar.
Inoculations of non-wounded fruits (i.e., intact cuticle and epidermis) were performed in parallel with inoculations of wounded fruits, which were made by the application of a 10 μL droplet of inoculum to a wound created by breaching the epidermis with a 22 gauge needle to generate a small hole to a depth of 2 mm. Lesion diameter (mm) was recorded three days after inoculation and incubation of the fruits in the humidified containers at room temperature (25 ± 1°C). Disease severity for each genotype was calculated as average lesion diameter × disease incidence (proportion of fruit with lesions greater than 3 mm). Standard cultivars (‘Ross’ or ‘Carson’) that are susceptible to brown rot were included each week, depending on their maturity and availability, as a positive control for each week’s disease assay.
BR susceptibility was categorized based on data of disease severity in the progenies and the empirical field observations and documented information of relative disease exhibition for non-inoculated breeding selections and cultivars. Thus, five categories were created, with 1 indicating resistance and 5 indicating high susceptibility (Table 1). For old cultivars and breeding selections, because they were not considered in the inoculation experiment, values for BR and factors Year and (partially) for Treatment were lost; and recorded as missing values (i.e. not available, NA).
| Category | Value | Average Lesion Diameter (mm) |
|---|---|---|
| Highly resistant | 1 | 0–5 |
| Moderately resistant | 2 | 5–20 |
| Susceptible | 3 | 20–30 |
| Moderately susceptible | 4 | 30–50 |
| Highly susceptible | 5 | >50 |
An ordinal logistic model with a logit link function (Agresti 2013) was used to calculate EBVs taken as the random effects term of a logistic generalized linear mixed model. Ordered polytomous modeling was undertaken for each possible score (on the scale) at which a threshold could be used to divide genotypes into one of the five categories (C): highly resistant, moderately resistant, susceptible, moderately susceptible and highly susceptible. The probability
Thus, it is possible to fit the reparameterized Bayesian additive genetic model proposed by Damgaard (2007) [based on the additive genetic model proposed by Bulmer (1980)], which derives from the univariate case of the Bayes rule:
Pr(θ/y) ∝ Pr(y|θ)Pr(θ), in which, we substituted θ to represent the joint distribution of β, the additive effects (a) and the square root of the additive genetic variance
The variance matrix of a is given as
Through the application of Markov chain Monte Carlo methods (Hastings 1970, Metropolis et al. 1953) the posterior sample distribution of a and
BOAM was evaluated using JAGS (Plummer 2003) version 3.4.0, using the library rjags 3-13 (Plummer and Stukalov 2014) to interface with R version 3.1.0 (R Development Core Team 2014). Since many of the genotypes were not evaluated over all of the years considered in the study, many individuals exhibited missing values for the factor year (which could take values of 1, 2 or 3). Values for year for these individuals were imputed through the dcat() function in JAGS, giving equal probabilities (1/3) to each one of the possible outcomes (1, 2 or 3). BOAM was fitted by running three independent chains for a total of 170,000 iterations, and a burn-in of 20,000, per chain, and model parameters were saved every 30th iteration, in order to yield 5000 samples to draw inferences. The category 1 for treatment (non-wounded) and year (year 1 = 2007) was fixed as zero. The priors for γj and β were considered as normally distributed with mean zero and variance 100 [γj ~ N(0, 100) or βlk ~ N(0, 100)], for which γj with j = 1, 2, 3, 4, the constraint is that γ1 < γ2 < γ3 < γ4. Therefore, the initial values for γj and the slopes for treatment and year (βlk) were set as follows: γ1 = −0.5, γ2 = 0.0, γ3 = 0.5, γ4 = 1.0, β1k = NA, β2k = 0.1, βl1 = NA, βl2 = 0.1 and βl3 = −0.1.
For
Parameters γj, β,
To the categorization presented in Table 1, an additional criterion related to the breeding value per se for peach improvement was added; that is to say, the score based on the category and given to a genotype also included an empirical judgment of the likely use of the selection for peach genetic improvement. Since for old cultivars and breeding selections inoculation experiment was not available, 1034 data points, were considered for the analysis, of which 4% were missing data and imputed within the BOAM procedure. Considering the Year and Treatment combinations showed a clearly distinct behavior among combinations (Fig. 2), which provides support to the approach of considering simultaneously these factors into a logistic model.

Trellis plot showing the density distribution of the data according to Year-Treatment combinations. On axis X the distinct degrees of BR susceptibility as described in Table 1 are shown.
The complete analysis required 12 hours, 5.2 GB RAM and 3.6 GB ROM on a desktop computer with a 2.8 GHz quad-core microprocessor.
The posterior mean, also known as the empirical mean, and the interval of the possible values for each parameter estimated through the model and with a respective prior for
| Estimated value for: | Half-Normal distributed prior | Gamma-distributed prior | Half-Cauchy-distributed prior | Uniformly distributed prior | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Empirical mean | Quantiles | Empirical mean | Quantiles | Empirical mean | Quantiles | Empirical mean | Quantiles | |||||
| 2.50% | 97.50% | 2.50% | 97.50% | 2.50% | 97.50% | 2.50% | 97.50% | |||||
| γ1 | −2.35 | −3.11 | −1.62 | −2.32 | −3.08 | −1.59 | −2.31 | −3.06 | −1.59 | −2.35 | −3.11 | −1.62 |
| γ2 | −0.66 | −1.41 | 0.06 | −0.63 | −1.37 | 0.09 | −0.62 | −1.36 | 0.09 | −0.66 | −1.41 | 0.06 |
| γ3 | 2.63 | 1.11 | 3.88 | 2.63 | 1.13 | 3.88 | 2.16 | 1.02 | 3.69 | 2.62 | 1.11 | 3.85 |
| γ4 | 2.14 | 1.01 | 3.69 | 2.15 | 0.99 | 3.70 | 2.63 | 1.15 | 3.85 | 2.14 | 0.99 | 3.77 |
| β1k0 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| β2k | −0.21 | −0.46 | 0.03 | −0.21 | −0.45 | 0.03 | −0.21 | −0.45 | 0.03 | −0.22 | −0.46 | 0.03 |
| βl1 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| βl2 | −0.05 | −0.44 | 0.34 | −0.04 | −0.44 | 0.35 | −0.05 | −0.44 | 0.34 | −0.05 | −0.45 | 0.34 |
| βl3 | −0.03 | −0.43 | 0.36 | −0.03 | −0.42 | 0.36 | −0.03 | −0.41 | 0.36 | −0.03 | −0.42 | 0.36 |
| h2 | 0.29 | 0.21 | 0.38 | 0.29 | 0.21 | 0.37 | 0.29 | 0.20 | 0.37 | 0.29 | 0.21 | 0.38 |
|
|
1.41 | 0.89 | 2.01 | 1.37 | 0.86 | 1.96 | 1.34 | 0.85 | 1.92 | 1.41 | 0.90 | 2.02 |
Empirical means and 2.50% and 97.50% quantiles for the parameters monitored in JAGS. These were calculated from the posterior distributions of each parameter.

Trace and density plots for heritability of BR susceptibility for each prior used to model the additive genetic variance.
The deviance values (including their penalization) for DIC for each BOAM using each prior are shown in Table 3, in which the Gamma-distributed prior showed the best fit. Interestingly, the uniform-distributed prior showed the least fit, but also the least penalty among models; hence, considering the penalized DIC value, the uniformly-distributed prior would be ranked second, followed by the Half-normal and Half-Cauchy prior. The differences between the DIC among models are shown in Table 4. Thus, the Gamma-distributed prior showed the best fit considering mean DIC but considering the pair-wise differences among models taking into account the penalized deviance, the uniformly distributed prior showed the best fit. However the difference is less than 2 units with the Gamma-distributed prior, and around 3 units with respect to the half-normal-distributed prior.
| Prior | Mean deviance | Penalty | Penalized deviance |
|---|---|---|---|
| Gamma | 4186 | 82.68 | 4248 |
| Half-normal | 4194 | 75.93 | 4270 |
| Half-Cauchy | 4196 | 93.98 | 4289 |
| Uniform | 4198 | 68.77 | 4266 |
For each model DIC were calculated using each prior for the additive genetic variance. Their locations correspond to the best-fitted model (top) to the least-fitted (bottom).
| Half-Cauchy | Gamma | Half-Normal | Uniform | |
|---|---|---|---|---|
| Half-Cauchy | − | 20.80438 | 19.44639 | 22.51744 |
| 15.16039 | 18.60957 | 16.4013 | ||
| Gamma | −20.80438 | – | −1.357996 | 1.713055 |
| 15.16039 | 17.86342 | 17.0784 | ||
| Half-Normal | −19.44639 | 1.357996 | – | 3.071051 |
| 18.60957 | 17.86342 | 15.54499 | ||
| Uniform | −22.51744 | −1.713055 | −3.071051 | – |
| 16.4013 | 17.0784 | 15.54499 |
The differences in DIC and corresponding standard errors among BOAM applying a distinct prior. Thus, the difference between the Half-Cauchy and Gamma-distributed prior with respect to DIC is 20.80438 and the sample standard error of this difference is 15.16039. These values show that the model using a Gamma-distributed prior for the additive genetic variance fits better than does a Half-Cauchy-distributed prior, which is observed in the difference of this same comparison but in the direction Gamma vs. Half-Cauchy, in which the DIC’s difference is negative, in favor for the Gamma-distributed prior.
The values for the EBVs ranged from −3.2546004 to 4.81101782 (Table 5), with the lowest values indicating resistance to BR and the highest indicating total susceptibility. The EBVs calculated through BOAM using distinct priors showed total correlation for the genotypes considered here, independently of the prior used. In contrast the EBVs estimated through REML showed lower values of correlation with the EBVs estimated through BOAM (Fig. 4).
| Ranking | Genotype | Half-Normal | Gamma | Half-Cauchy | Uniform |
|---|---|---|---|---|---|
| 2 | Bolinha | −3.254600391 | −3.178712853 | −3.171238835 | −3.23762941 |
| 3 | 01,9-271 | −2.712693236 | −2.680472621 | −2.664983005 | −2.709349014 |
| 6 | Nonpareil-BF | −2.396573347 | −2.387498349 | −2.381702153 | −2.40188797 |
| 7 | Orange Cling | −2.396573347 | −2.387498349 | −2.381702153 | −2.40188797 |
| 8 | 01,9-035 | −2.291572396 | −2.265968122 | −2.254664605 | −2.296784474 |
| 15 | 01,9-082 | −2.276717584 | −2.254075204 | −2.244794489 | −2.282397147 |
| 26 | 02,2-255 | −2.239161688 | −2.222356864 | −2.210881871 | −2.256686101 |
| 27 | 02,2-258 | −2.239161688 | −2.222356864 | −2.210881871 | −2.256686101 |
| 28 | 02,7-086 | −2.247504491 | −2.213871168 | −2.211154439 | −2.247187379 |
| 29 | 02,7-088 | −2.247504491 | −2.213871168 | −2.211154439 | −2.247187379 |
| 212 | Dix-6-6 | 3.111762993 | 3.109898726 | 3.081505407 | 3.106417645 |
| 213 | Dix-8 | 3.111762993 | 3.109898726 | 3.081505407 | 3.106417645 |
| 214 | Everts | 3.399790384 | 3.359438477 | 3.353500054 | 3.397536024 |
| 215 | Jungerman | 3.399790384 | 3.359438477 | 3.353500054 | 3.397536024 |
| 216 | Dix-9 | 3.785353243 | 3.760980686 | 3.7623027 | 3.781094126 |
| 217 | Dixon | 3.785353243 | 3.760980686 | 3.7623027 | 3.781094126 |
| 218 | Dix-22A-5 | 4.797976669 | 4.747679292 | 4.718376925 | 4.793894615 |
| 219 | Dix-5A-1 | 4.797976669 | 4.747679292 | 4.718376925 | 4.793894615 |
| 220 | D-30-3E | 4.811017817 | 4.754630563 | 4.736022267 | 4.791423132 |
| 221 | D25-9E | 4.811017817 | 4.754630563 | 4.736022267 | 4.791423132 |

Scatter plot of matrices (SPLOM) showing the correlation of EBVs calculated through BOAM for each prior and through REML. Scatter plots below the diagonal show the linear regression that fits for the two sets of EBVs, shown on the diagonal frequency histograms, while Pearson correlation (with pairwise deletion) values are shown above the diagonal, generated though using the function pairs.panels of the package psych version 1.4.4 (Ravelle 2014) for R version 3.1.0. EBVs_HNo: EBVs calculated with an additive genetic variance with a Half-Normal-distributed prior, EBVs_Gam: EBVs calculated with an additive genetic variance with a Gamma-distributed-prior, EBVs_HCa = EBVs calculated with an additive genetic variance with a Half-Cauchy-distributed-prior and EBVs_Uni: EBVs calculated with additive genetic variance with an uniformly distributed prior, EBVs_REML: EBVs calculated through REML using a BR severity index for wounded and non-wounded fruits of Pop-BR-1 and PopBR-2, EBVs_REMW: EBVs calculated through REML using a BR severity index for wounded fruits only from PopBR-1 and PopBR-2, REMN: EBVs calculated through REML using a BR severity index for non-wounded fruits only from PopBR1 and PopBR-2.
The results of this study clearly show that the information provided by the dataset and the strategy followed are robust. This is supported by the null influence of the priors used for the estimation of genetic parameters such as the additive genetic variance
Although the original analysis required 12 hours to be performed due to the number of iterations solicited (170,000), for this dataset, this amount of time can be severely reduced using 7000 iteration only, using a burn-in period of 2000 iterations and avoiding the application of thinning, in order to keep 5000 samples for parameter inferences.
Because of the advancements in personal computational power, it has become feasible to perform statistical analysis based on MCMC such as the one presented here, while previously it was problematic (Damgaard 2007). It is expected that refinement of existing algorithms and development of new ones will enable researchers and applied breeders to fit more complicated models to more accurately estimate genetic parameters and additional terms such as interactions. In addition, more efficient informatics samplers will enhance the speed at which results are produced. Examples can be seen in the development of Stan (Hoffman and Gelman 2014, Stan Development Team 2015) and in new approaches not based on sampling, such as Integrated Nested Laplace Approximation (INLA, see Holand et al. 2013) and the development of faster programming languages such as Julia (Bezanson et al. 2012).
The absence of difference among the estimations of
It is commonly known that in order to get reliable and robust estimates through logistic regression, a complete dataset (i.e. no missing data in variables and factor levels) is preferred overall to avoid breaking the assumption of random missingness which affects the pattern of risk factors observed in a whole dataset (Jewell 2004). Through the implementation of an imputation of factor levels for Year, it was possible to get robust estimations for genetic parameters regardless of missing data and priors applied. Such an imputation of factor levels enabled us to take into account the available information for members of the pedigree that were not evaluated in the study. This is the case of commercial cultivars and old breeding selections, for which information can be obtained, for instance, in documented records of resistance in cultivar catalogs and extension reports or passport information from germplasm repositories.
The application of the Bayesian ordinal animal model on the study of brown rot resistance in peachBR resistance in peach is the result of interactions among multiple structural and biochemical components; in the fruit epidermis and flesh which show large environmental effects (Cantoni et al. 1996, Gradziel et al. 2003, Gradziel and Wang 1993). Many of the components and their interactions can be measured, though the amount of information provided per component is restricted to the proportional effect of the component in the exhibition of BR resistance (Gradziel et al. 1998). However, the formation of categories involves a subjective component; in this case, the categorization is well-based on the understanding of the morphological and biochemical components of peach fruit involved in the exhibition of BR infection. Although, the gene is the inheritance unit, the phenotype (the trait itself) continues being the reference unit for breeding. Thus, the use of categories based on breeder expertise is a summarization of all the processes involved in BR resistance exhibition; processes that often do not require genetic dissection for breeding to be performed; although any additional information from dissections is highly desirable, particularly for marker-assisted selection. The information provided by the application of BOAM serves as a first support for decision-making in the breeding program to assist selection and design of crosses. Also, information based on BOAM can be useful to assist genetic dissection of traits by selecting those individuals with extreme EBVs and known lineages.
The utilization of Bayesian inference techniques through an ordinal logistic model has enabled us to obtain the first estimation for h2 for BR resistance within a pedigree of peach, which reveals that the influence of the genetic component involved in the exhibition of this trait is certainly not marginal, and there is still sufficient genetic variation to support advancements for the genetic improvement of peach, as has been demonstrated by Gradziel (see Gradziel et al. 2003 and Gradziel 2012) through the application of recurrent back-crosses. The h2 range from 0.21 to 0.38 is realistic in the light of the empirical experience gained in the breeding program.
Typically, the study of genetic traits begins with estimating genetic parameters such as additive genetic variance, genetic effects beyond additivity (dominance and epistasis), heritability, genetic correlations and breeding values. Previous estimates of genetic parameters in peach have been limited to the study of bi-parental populations and, in some cases, half-sib families (Hansche 1986, 1990, Hansche and Boynton 1986, Hansche et al. 1972). Estimating genetic parameters, including EBVs, within a pedigree has occurred only in the studies led by de Souza and Byrne (de Souza et al. 1998a, 1998b, 2000), where REML was applied to estimate variance components. This scenario agrees with the assessment done by Piepho et al. (2008) for the case of annual crops, in which the estimation of EBVs through BLUP is not as common as it is for animal breeding, regardless of the power of the methods. For annual crops, selection is not usually based on the individual genotype performance, as it is in animal (principally livestock) breeding. In this regard, fruit tree crop breeding has more similitudes to animal breeding (e.g. selection on individual genotypes, pedigree records, repeated measures, limited progeny size, exhibition of long juvenility), and thus the extension of the animal model is applicable.
Our estimation of h2 is in general agreement with those reported for other fungal disease resistance analyses in peach, such as peach leaf curl (Taphrina deformans (Berk.) Tul.] which was estimated as 0.34 ± 0.19 (Ritchie and Werner 1981), suggesting that the resistance to these fungal infections tends to be inherited quantitatively. Quantitative control is also supported by the mapped QTLs for BR resistance (Martínez-García et al. 2013) and peach leaf curl (Viruel et al. 1998). For another fungal infection, peach canker [Leucostoma persoonii (Nitschke) Höhn.], h2 was estimated as 0.72 (Chang et al. 1991), which suggests that for this particular disease the inheritance tends to be Mendelian (one or two genes) rather than quantitative (more than three genes).
Since the ordinal logistic model can be considered as an extension of the general linear model to ordinal categorical data, and hence of the of binary logistic model (Menard 2002), changes in the scale (assuming appropriate sample size, null or minimum missing data, same ordinal usage for the categories, and independence among the dependent variable choices) would be minimal for the ranking of the EBVs due to the estimation of
Previous analyses applying the Average Information Algorithm on Restricted Maximum Likelihood (REML, Gilmour et al. 1995) on the severity index data (a continuous response variable) previously obtained for PopBR-1 and PopBR-2 only, yielded more optimistic h2 values. That analysis used data from both treatments (wounded and non-wounded fruits). The estimations (in gamma scale = variance component ratios) were
The EBVs yielded through BOAM, independently of the prior applied, are consistent with the empirical experience. Thus, the progenies from the families PopBR-1 and PopBR-2 show the expected segregation of the trait. Similarly the values for breeding lines and plant introductions were in accordance with their average performance across the years, and in the case of commercial cultivars, the values and ranks are in accordance with their documented descriptions. Even though the values of h2 yielded through REML can be considered as similar to the range reported here, the main reason for seeking out an alternative strategy for estimation of h2 and EBVs for BR resistance is the unrealistic ranking of the EBVs resulting from REML using a BR severity index. These issues can be seen in Fig. 4, where the deficient correlation with the EBVs estimated through BOAM using distinct priors is evident. Thus, the genotype ‘Bolinha’, recognized as the source of BR resistance in peach, was ranked 155th out of 221 and ‘Dixon’, known for its high susceptibility to BR, was ranked 102st out of 221, while for BOAM the rankings of these cultivars were 2nd and 217th out of 221, respectively. Similar outcomes have been reported previously; EBVs calculated through a mixed linear mixed model were not co-linear with EBVs estimated through an ordinal mixed model, which led to the conclusion that, when estimating EBVs for traits with ordinal responses, an ordinal mixed model is preferable (Wilson et al. 2013). This last conclusion supports the performance of BOAM, since this method is also realistic and considers the ordinal nature of BR resistance and other traits.
In addition, BOAM can be extended for more accurate estimations of EBVs, which can be approached through the addition of other ordinal covariates, such as fruit maturity stage, agronomic treatments and regimes, postharvest handling treatments such as time of cold conservation, or type of inoculum/inoculation. However, note that since the BOAM is based on iterative methods, the amount of time and computational power to fit the model(s) and achieve convergence can increase considerably. Also, model selection is advised, in order to identify the set of models holding desirable statistical characteristics (precision and ease of handling) as well as realistic predictions.
Perspective of the application of BOAM in the breeding programEffective selection for BR resistance in peach is becoming increasingly important for breeders and ultimately for growers and consumers as the use of chemical fungicides is reduced or eliminated. While important advances have been achieved during the last twenty years in the understanding of the nature of the pathogen(s) and the response of peach to infection (see Oliveira et al. 2016), the efficient selection for BR resistance remains challenging. Factors affecting efficacy include the few documented sources of resistance such as ‘Bolinha’, the harvest of fruit at the full-ripe stage which maximizes quality but also vulnerability, and the strong selection against enzymatic browning of fruit tissue despite its association with Bolinha-type resistance (Gradziel et al. 1998). Consequently, the introgression of novel sources of resistance from almond and related species has been pursued targeting genetic gain of desirable BR resistance alleles simultaneously from both peach and exotic sources through recurrent backcrossing. The development of BOAM provides the required statistical tools for: a) the combination of phenotypic data from evaluations of wounded and non-wounded fruit which appear to represent distinct biological responses and so potential BR resistance alleles, and b) the phenotypic summarization of BR resistance expression through the use of a breeding relevant ordinal categorization. Because several mechanisms are known to be involved in BR resistance, including phenolic concentration, thickness of epidermis components, and flesh texture, selection should be simultaneously based on all known as well as unknown components, which according to our analysis are significant, having heritability ranging from 0.21 to 0.38. Thus, the application of BOAM allows the breeder to summarize the phenomenon into categories, and thus subsequently consolidate available phenotypic information through defining individuals with high genetic merit (supported by their EBVs) to be further evaluated and potentially used as parents in further optimized breeding schemes. In the effort to develop peach cultivars combining resistance from both almond and peach, results yielded by BOAM propose crosses with a higher probability, as expressed by their EBVs, of recovering progenies with combined resistance. For example, promising parental genotypes would include ‘01,9-271’, ‘01,9-035’ or ‘01,9-082’ from PopBR-1 having the resistance from almond, combined with ‘02,2-255’, ‘02,2-258’, ‘02,7-086’, or ‘02,7-088’ from PopBR-2 having resistance from peach. Bidirectional crosses would be recommended to determine whether there are maternal effects for this trait.
Three important factors have to be considered when breeding for disease resistance (Singh et al. 2002). The first consideration is the nature of the pathogenic agent, including the diversity of virulence. The second is knowledge of the availability, diversity and types of genetic resistance within the breeding program, as well as within the species and close relatives. The third is the handling, developing and improving of screening methods and phenotyping, including accurate selection of the appropriate environment for exhibition of resistance to allow its accurate tracking. The breeder plays a major role in addressing these factors, for the optimization of resources, which is fundamental to a successful breeding program. The estimation of h2 and EBVs is a tool for understanding and manipulating the exploitable genetic components available within breeding germplasm. Its use does not exclude additional strategies and tools and, in fact, can complement them. For example, the EBVs estimated here may be used to expand and support the results generated through QTL mapping, since the joining of the genotypes or haplotypes of the molecular markers associated with the trait may be related to the breeding value, and thus, imply the most probable genetic/gametic phase for the selection of parents. This approach is pursued through the Pedigree Based Analysis under the Bayesian Framework (Bink et al. 2002, 2004, 2008, 2012, 2014), which enables breeders and researchers to map QTLs while providing prior knowledge about the number of QTLs that may be involved in the exhibition of the trait, and which thus yields genome positions and the denominated Genome-wide Estimated Breeding Values.
Here, a strategy based on Bayesian inference and breeder phenotypic information was developed and applied to estimate genetic parameters such as h2 and EBVs to assist in selecting parents for BR resistance. BOAM was successfully applied on a pedigree possessing two sources of resistance, one from peach and the other from almond, and with historical information from several well-known commercial cultivars, breeding selections, as well as two segregating populations. The results of BOAM proved more realistic than those yielded through analysis of a severity index through REML. These findings have been valuable for the genetic improvement for BR resistance by optimizing the introgression of both the peach and the almond alleles. This approach should prove useful for other species in which the traits of interest are ordinal and a pedigree structure is available. The continuous refinement of procedures based on Bayesian inference will allow future access to more robust quantitative genetic methods for fruit crop breeding, and we expect that application of procedures, such as those presented here, will be extended to incorporate information from QTLs, haplotypes and molecular markers, as they become feasible through the addition of appropriate vectors and matrices as terms of the model.
The authors gratefully acknowledge CONACYT-UCMEXUS for providing PhD fellowship support to Jonathan Fresnedo Ramírez, and to Palma Lower, writing specialist at UC Davis, for her valuable comments and corrections during the redaction of the manuscript. The technical assistance of Tatiana Roubtsova, Eva Gutierrez, Alex De Beaumont-Felt, Emilie Trinh, Timothy Weerts, Rachel Giffin, Thanh Le, Karl Lund, Mary Ann Thorpe, and field assistants to the UC Davis Peach Breeding Program are gratefully acknowledged.