2018 Volume 68 Issue 2 Pages 248-257
2018 Volume 68 Issue 2 Pages 248-257
Genomic selection is attracting attention in the field of crop breeding. To apply genomic selection effectively for autogamous (self-pollinating) crops, an efficient outcross system is desired. Since dominant male sterility is a powerful tool for easy and successive outcross of autogamous crops, we developed transgenic dominant male sterile rice (Oryza sativa L.) using the barnase gene that is expressed by the tapetum-specific promoter BoA9. Barnase-induced male sterile rice No. 10 (BMS10) was selected for its stable male sterility and normal growth characteristics. The BMS10 flowering habits, including heading date, flowering date, and daily flowering time of BMS10 tended to be delayed compared to wild type. When BMS10 and wild type were placed side-by-side and crossed under an open-pollinating condition, the seed-setting rate was <1.5%. When the clipping method was used to avoid the influence of late flowering habits, the seed-setting rate of BMS10 increased to a maximum of 86.4%. Although flowering synchronicity should be improved to increase the seed-setting rate, our results showed that this system can produce stable transgenic male sterility with normal female fertility in rice. The transgenic male sterile rice would promote a genomic selection-based breeding system in rice.
Genomic selection (Meuwissen et al. 2001) has been applied to dairy cattle breeding in the last decade, and cumulative selection effects have been recently reported (García-Ruiz et al. 2016). Genotyping costs are very reasonable, especially in the breeding of large-size livestock animals, compared with the cost of foraging for a parental candidate population that could take years. Recently, the cost of genome-wide genotyping was reduced drastically and genomic selection attracted attention as a new method of plant breeding (Heffner et al. 2009, Jannink et al. 2010, Lorenz et al. 2011). However, genomic selection is based on repeated outcross propagation (Meuwissen et al. 2001), making it difficult to apply continuous genomic selection directly in the breeding programs of autogamous (self-pollinating) crops, such as rice (Oryza sativa L.), wheat (Triticum aestivum L.), and soybean (Glycine max L.). The conventional breeding programs of autogamous crops have taken time over the last decade to evaluate crossings, genetic fixation by selfing, and phenotypic selection. To bring out the power of cumulative genomic selection, breeders are required to conduct artificial crossings at every generation. When we aim to apply genomic selection to breeding programs of autogamous crops, an efficient outcross system is essential.
Outcross using male sterility has been proposed and practiced in autogamous crop breeding for a long time (Brim and Stuber 1973, Doggett and Eberhart 1968, Fujimaki 1980). However, these outcross systems were not a major breeding method in autogamous crops, because they have two main problems that prevent large-scale and common breeding. The first is that the phenotype of genetic male sterility in these systems is recessive. These male sterile plants do not appear in the F1 generation when genetic male sterile plants are crossed with wild type, and theoretically, one-fourth of plants would show male sterility in the F2 generation. This segregation ratio of male sterile plants has become an obstacle to building an efficient outcross system. Another problem is that so far these systems do not have completely linked phenotypic markers to discriminate between male sterile and male fertile individuals (Brim and Stuber 1973, Doggett and Eberhart 1968, Fujimaki 1980). Breeders should distinguish them in the population during the flowering stage, despite this being the busiest period in a year. Falk et al. (1981) and Falk and Kasha (1982) used a shrunken endosperm (sex1) phenotype of seeds as a selection marker to easily discriminate between male sterile and male fertile plants. However, this linkage between sex1 and male sterility is sometimes broken by genetic recombination. To solve these problems, Tanaka (2010) proposed a selectable dominant male sterile system that uses positive and negative selection markers that can easily select male sterile or male fertile plants. In addition, the final products of this system do not contain the transgene (null segregants) because the true-bred cultivars must not be male sterile. Competent authorities have decided that null segregants have been out of the regulation of genetically modified crops in the United States (https://www.aphis.usda.gov/aphis/ourfocus/biotechnology/am-i-regulated/Regulated_Article_Letters_of_Inquiry) and Argentina (Whelan and Lema 2015).
Mariani et al. (1990) generated dominant male sterility for rapeseed (Brassica napus L.) and tobacco (Nicotiana tabacum L.) using the barnase (ribonuclease from Bacillus amyloliquefaciens, Acc. No. M14442, EC 3.1.27, Paddon and Hartley 1985) gene that is expressed by the tapetum-specific promoter TA29 in tobacco. Subsequently, dominant male sterile plants were produced in many plant species such as wheat (De Block et al. 1997), oilseed mustard (Brassica juncea L.) (Jagannath et al. 2001), maize (Zea mays L.) (Sun et al. 2008), eggplant (Solanum melongena L.) (Cao et al. 2010), pine (Pinus radiate L.) and eucalypts (Eucalyptus spp.) (Zhang et al. 2012), and pelargonium [Pelargonium zonale (L.) L’Hér. ex Aiton] (García-Sogo et al. 2012). Lu et al. (2000) reported on dominant male sterile rice using the barnase gene that was expressed by the anther-specific promoter. However, this male sterile rice has not been analyzed in detail, and an application for this rice has still not been found. The tapetum plays an important role in the development of male gametophytes, and premature destruction of the tapetum that is known to cause male sterility (Scott et al. 1991). Therefore, most of the male sterile crop species described above were induced by cytotoxic genes such as barnase under the control of tapetum-specific promoters.
In this study, we generated transgenic dominant male sterile rice using the barnase gene expressed by the tapetum-specific promoter BoA9 from broccoli (Brassica oleracea L.) (Konagaya et al. 2008) and the barstar gene driven by the CaMV35S promoter to develop an efficient outcross system in rice. The leaky expression of the barnase gene in vegetative tissue often led to abnormalities in vegetative morphology, poor female fertility, low seed germination frequencies, and/or distortion in segregation ratios of transgenes (Jagannath et al. 2001, Lannenpaa et al. 2005, Wei et al. 2007). To avoid the harmful effect of barnase, barstar, which inhibits barnase activity by generating a complex with barnase, was often co-introduced under a CaMV35S promoter (Gardner et al. 2009, Wei et al. 2007). Because the CaMV35S promoter does not work well enough in the tapetum, this promoter is very useful to avoid the harmful effect of barnase on vegetative tissue (Gardner et al. 2009, van der Meer et al. 1992). Plants with the barstar expression under constitutive promoters showed normal growth and morphology, the same as non-transgenic plants (Gardner et al. 2009, Wei et al. 2007). We then evaluated flowering habits, which greatly influences outcross efficiency. Our results demonstrated that transgenic dominant male sterility of rice could provide a useful tool for an efficient outcross system that would promote a genomic selection-based breeding system.
A plasmid vector pBoA9::GUS (Fig. 1) was constructed based on pIG121-Hm (Acc. No. AB489142, Ohta et al. 1990). The fragment containing the BoA9 promoter and GUS gene was cut by SbfI and SacI from the BoA9::GUS vector (Konagaya et al. 2008). The CaMV35S promoter and CAT1-GUS gene of pIG121-Hm were replaced with a fragment of the BoA9 promoter and GUS gene by the SbfI/SacI site.

Structure of the T-DNA region in the binary vector pBoA9::GUS and pHA9Bn-Bs. P-nos: promoter of the nopaline synthase gene (NOS); NPTII: neomycin phosphotransferase II gene; T-nos: terminator of the NOS gene; P-BoA9: tapetum-specific promoter from broccoli; GUS: β-Glucuronidase gene; P-35S: the 35S promoter from Cauliflower mosaic virus; HPT: hygromycin phosphotransferase gene; Barnase: barnase gene encoding the ribonuclease from B. amyloliquefaciens; intron: the first CAT1 intron of castor bean; Barstar: barstar gene from B. amyloliquefaciens encoding inhibitor of barnase; LB and RB: left and right border of the T-DNA. The thick line indicates the probe region for the Southern blot analysis. PmeI, ApaI, SbfI, AscI, SacI, XhoI, and BamHI indicate the restriction site of individual restriction enzymes.
A plasmid vector pHA9Bn-Bs (Fig. 1) was constructed based on pIG121-Hm. Primer sequences used for plasmid construction are shown in Supplemental Table 1. The plasmid involved three gene cassettes: (1) the barnase gene-expressing cassette, (2) the barstar gene-expressing cassette, and (3) the resistant to hygromycin cassette.
Agrobacterium-mediated transformation of rice (cv. ‘Nipponbare’) was performed as previously described (Toki et al. 2006).
Plant materials and growth conditionsNon-transgenic rice (cv. ‘Nipponbare’ and ‘Tachiaoba’) and transgenic rice were grown in commercial soil (Bonsoru No.1, Sumitomo Chemical, Tokyo, Japan) in a growth chamber under white fluorescent light (230 μmol photons m−2 s−1) with a 10 h light (27°C)/14 h dark (25°C) cycle and a concentration of 600 ppm CO2 (biotron breeding system condition) (Tanaka et al. 2016), or a closed greenhouse programmed for day/night temperatures of 28°C/24°C, or the climate-following closed greenhouse under a natural photoperiod. We selected a transgenic male sterile rice line named barnase-induced male sterile rice (BMS) 10 and used it for the following crossing experiments. The progeny derived from a crossing between BMS10 and ‘Tachiaoba’ were germinated and grown in Murashige and Skoog (1962) medium in a growth chamber under continuous white fluorescent light at 27°C.
Histochemical GUS assayA GUS assay was performed following the method of Konagaya et al. (2008) with minor modifications, followed by 5 mM potassium ferrocyanide and 5 mM potassium ferricyanide. The pollen was photographed on an MZ16FA stereomicroscope with an HL RC transmitted light base (Leica Microsystems, Switzerland). The pollen developmental stage was classified as described Jung et al. (2005)
Characterization of male sterility of T0 and T1 generation in BMSThe T0 and T1 generations in BMS were cultivated in the closed greenhouse. To observe pollen viability, mature anthers of BMS and wild type individuals were stained with Alexander’s solution (Alexander 1969) and photographed under a VANOX-T microscope. At least 60 anthers were examined for each plant. The BMS panicles were enclosed in bags just after heading to avoid crossing by other pollens, and the number of seeds were counted after harvesting.
Confirmation of female fertility of T0 generation in BMSThe T0 individuals of BMS as female parents were crossed with wild type as male parents. The already opened BMS spikelets were removed and the upper part of remaining BMS spikelets were cut off (clipping method). These BMS and wild type panicles were tied in a bundle and enclosed together in bags. The bags containing the panicles were then shaken hourly from 10:30 to 14:30 for 3 days after bagging.
Phenotyping and evaluation of flowering habits of BMST0 individuals were classified as one of five grades by the percentage of flowering spikelets against total spikelets (1: 0–10%, 2: 11–20%, 3: 21–30%, 4: 31–50%, 5: >50%) to select T0 individuals that showed a high frequency of flowering spikelets.
The flowering habits in the T1 generation of BMS were observed in more detail. The number of flowering spikelets in the T1 generation were counted hourly from 10:00 to 17:00 h each day during the 2 weeks after heading.
Crossings between BMS10 and ‘Nipponbare’ or ‘Tachiaoba’Three different cross-pollination methods were used in the following examinations. In the first and second experiments, T1 individuals of BMS10 were crossed with ‘Nipponbare’. BMS10 and ‘Nipponbare’ panicles were tied in a bundle and enclosed in bags in the first experiment. Then, we evaluated the seed-setting rate in the near-natural culture condition in the second experiment. BMS10 and ‘Nipponbare’ were laid side-by-side with intervals of about 20 cm and cultivated in the closed greenhouse. In the third experiment, ‘Tachiaoba’ was used as the pollen parent to demonstrate hybrid productive efficacy. The clipping method was used. Clipped BMS10 and ‘Tachiaoba’ panicles were tied in a bundle and enclosed in bags. The panicles were shaken hourly from 10:30 to 14:30 for 3 days after bagging in the first and third experiments, and every day during the 2 weeks after heading in the second experiment. Hybridity between BMS10 and ‘Tachiaoba’ was confirmed by a simple sequence repeat (SSR) marker.
Detection of inserted genes in BMSGenomic DNA was isolated from the leaves of BMS seedlings in T0 and T1 individuals following the method of Edwards et al. (1991) with phenol-chloroform extraction. To detect the inserted genes in T0 and T1 individuals, PCR was performed using a thermal cycler (Gene Amp PCR system 9700, Applied Biosystems, USA). Amplification was performed for an initial denaturation at 94°C for 2 min, 30 cycles at 94°C for 30 s, 60°C for 30 s, 72°C for 1 min, and a final extension at 72°C for 5 min with the primer sets HPT-F01 and HPT-R01 for HPT and Barnase-F02 and Barnase-R02 for barnase. Primer sequences are shown in Supplemental Table 1. The amplified DNAs were electrophoresed in 1.5% agarose gel.
Southern blot analysis of T0 and T1 generation in BMS10Genomic DNA was isolated from the leaves of ‘Nipponbare’ and T0 and T1 BMS individuals using ISOPLANT II (Nippon gene, Tokyo, Japan). Ten micrograms of genomic DNA was digested with ApaI or XbaI. The digested DNAs were separated in 1.0% agarose gel and blotted onto a Hybond N+ membrane (GE Healthcare, Piscataway, NJ, USA). A 529 bp region of the HPT gene was labeled using the DIG PCR Labeling Mix (Roche Applied Science, Penzberg, Germany). PCR for probe labeling was carried out using the primer set HPT-F01 and HPT-R01, the same as the detection of the inserted gene. The hybridization signals were detected using the DIG luminescent detection kit (Roche Applied Science).
Parentage test of the progenies of BMS10 by SSR markerGenomic DNAs were isolated from the leaves of ‘Nipponbare’, ‘Tachiaoba’ and F1 individuals derived from a crossing between BMS10 individuals (T1) and ‘Tachiaoba’. The genomic DNA isolation method and PCR condition were the same as that described in the section “Detection of inserted genes in BMS”. The polymorphic SSR marker RM5926 (McCouch et al. 2002) was used. Amplified products were separated by 3.0% agarose gel electrophoresis.
Tissue specificity of the BoA9 promoter in rice was examined by a histochemical GUS analysis. The results showed that the BoA9 promoter worked weekly only in the anther, mainly from the meiocyte to the tetrad stage (Fig. 2A, 2B). GUS activity was not observed in leaves, stems, or roots (data not shown). These results are similar to a study that used the BoA9 promoter in Arabidopsis (Konagaya et al. 2008).

Histochemical GUS analysis of T0 rice transformed pBoA9:: GUS. (A) Spatial and temporal expression pattern of the BoA9::GUS fusion product in spikelets at various developmental stage of pollen. Scale bars = 1 mm. (B) Magnification of the anther views in each stage. (a) Meiocyte, (b) Microspore. Scale bars = 0.5 mm.
Twelve independent transgenic T0 individuals were produced and the barnase gene and HPT gene were detected in all transgenic individuals by PCR (data not shown). Five of them showed abnormal or inferior growth and were eliminated, and the remaining seven individuals were used in the following experiments. To assess pollen productivity and viability, the anthers of BMS and wild type individuals were stained with Alexander’s solution. Although wild type individuals produced pollen grains, no pollen grains were observed in the anthers of any transgenic individuals under the same growth condition as the wild type (Fig. 3). The panicles of transgenic individuals were enclosed in bags just after heading to confirm male sterility; no BMS individuals produced seeds, although wild type showed a 96.2% seed-setting rate (Table 1). These results indicated that all examined T0 individuals exhibited male sterility. The rate of flowering spikelets in BMS individuals and wild type were also evaluated (Grades 1–5). Four BMS individuals (BMS3, 5, 6, and 10) showed a high frequency of Grade 4 or 5 (Table 1). Since BMS5 and BMS10 showed the same vegetative growth as that of wild type in the closed greenhouse, BMS5 and BMS10 were selected and used in the following experiments.
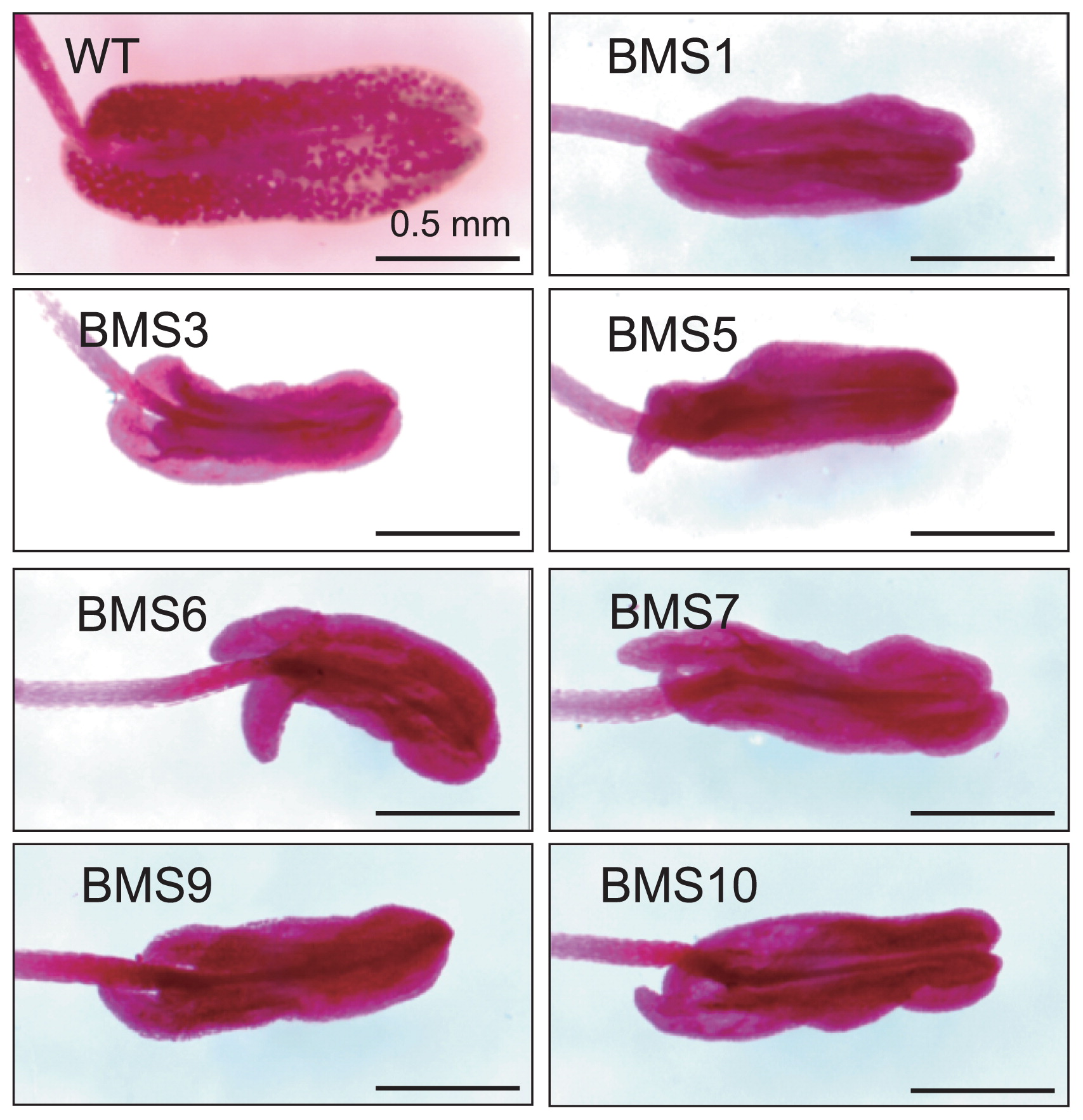
Anthers of wild type (WT) and BMS T0 individuals. Mature anthers of WT and BMS T0 individuals were stained with Alexander’s solution. Pollen grains were only observed in WT. All images are displayed with the same magnification. Scale bars = 0.5 mm.
| Line | No. of spikelets | No. of seeds | Seed setting rate (%) | Rate of flowering spikelets* (1–5) |
|---|---|---|---|---|
| BMS1 | 332 | 0 | 0.0 | 2 |
| BMS3 | 224 | 0 | 0.0 | 5 |
| BMS5 | 796 | 0 | 0.0 | 4 |
| BMS6 | 205 | 0 | 0.0 | 4 |
| BMS7 | 408 | 0 | 0.0 | 2 |
| BMS9 | 171 | 0 | 0.0 | 1 |
| BMS10 | 337 | 0 | 0.0 | 4 |
| WT | 105 | 101 | 96.2 | 5 |
WT: wild type.
T0 plants of BMS5 and BMS10 were crossed with a wild type to investigate female fertility. In the results of this cross experiment, 269 F1 seed from 427 spikelets and 526 seeds from 1,334 spikelets were produced in BMS5 and BMS10, respectively (Table 2). This result suggests that the female fertility of BMS5 and BMS10 was not affected by barnase protein toxicity. However, BMS5 produced three seeds from 47 spikelets by self-pollination. Since BMS10 showed complete male sterility and female fertility (Fig. 4A, 4B) and normal vegetative growth, BMS10 was selected and used in the following experiments.
| Cross combination | No. of spikelets | No. of seeds | Seed setting rate (%) |
|---|---|---|---|
| BMS5 (self-pollination)a | 47 | 3 | 6.4 |
| BMS5 × WTb | 427 | 269 | 63.0 |
| BMS10 (self-pollination)a | 128 | 0 | 0.0 |
| BMS10 × WTb | 1334 | 526c | 39.4 |
WT: wild type.

Phenotypes of BMS10 individuals. (A, B) Panicles from BMS10 (T0) after crossing with wild type (A) and self-pollination (B). Scale bars = 1 cm. (C) Wild type (‘Nipponbare’; left) and BMS10-24 and BMS10-33 (T1 individuals) (center and right). (D, E) Spikelet after removing the lemma and palea of the wild type (D) and BMS10-08 (T1) (E). Scale bars = 5 mm. (F, G) Anther of wild type (F) and BMS10-05 (T1) (G). Scale bars = 200 μm.
The number of T-DNA insertions was evaluated by a Southern blot analysis. Genomic DNA isolated from T0 and T1 progenies from BMS10 was digested with ApaI or XbaI and hybridized with an HPT probe (Fig. 1). A single fragment was detected in BMS10 T0 and T1 individuals by the probe as part of the hygromycin resistance gene (Fig. 5). Next, the segregation ratios of transgenic and non-transgenic rice in the T1 generation of BMS10 (BMS10 × wild type) were identified by PCR (Table 2). A total of 194 progenies, derived from crossings between BMS10 individuals (T0) and wild type, were tested; 84 individuals had transgenes (43.3%) and 110 individuals did not (56.7%). This result corresponded to the insertion of T-DNA into a single locus (χ2 = 3.48, P = 0.062 for a 1:1 segregation ratio). These results supported the consequence of the Southern blot analysis (Fig. 5).
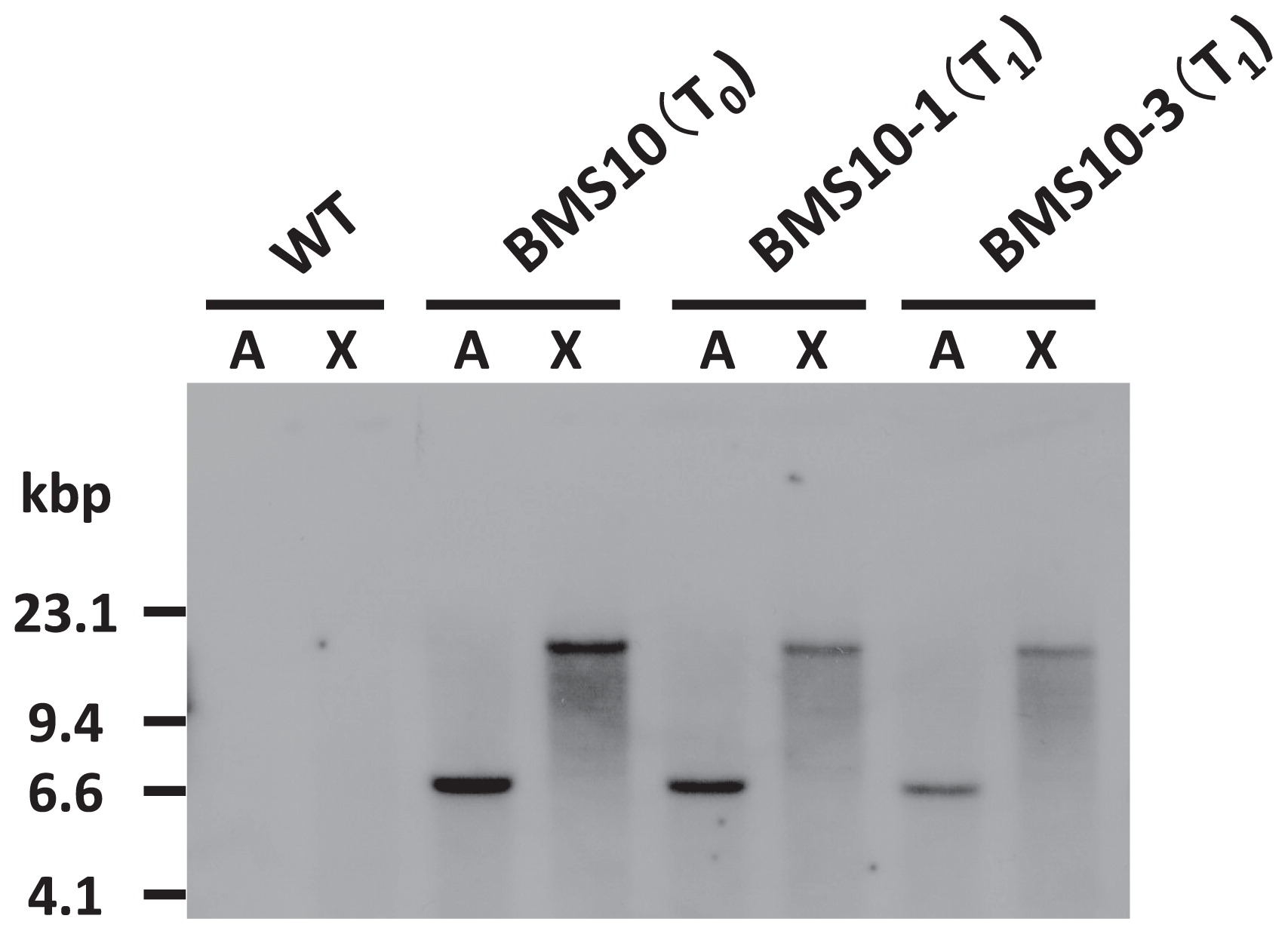
Southern blot analysis in BMS (T0 and T1 individuals). Ten μg of genomic DNA of wild type (WT), BMS10 (T0), BMS10-1 (T1), and BMS10-3 (T1) were digested with ApaI (A) or XbaI (X). The probe used in this experiment is shown in Fig. 1.
In the BMS10 T1 population, no abnormal phenotype was observed during the vegetative developmental stage (Fig. 4C). A total of 34 individual BMS10 T1 plants were assessed for male sterile stability. The degenerated anthers of BMS10 were compared with wild type (Fig. 4D, 4E), and no pollen grains were observed in the anther of any T1 BMS10 rice with transgenes (Fig. 4F, 4G). These results indicate that the male sterility phenotype was stable and inherited in the T1 generation.
Flowering habits of the T1 population of BMS10Heading dates of several wild type individuals were examined; however, inter-individual differences were hardly observed (data not shown). On the other hand, heading dates of BMS10 progenies with transgenes were later by 1–5 days than that of the wild type (Fig. 6). Although the flowering date of wild type began just after heading and most flowers opened within 3 days after heading, the flowering date of BMS10 progenies was delayed by 2–4 days and reached full bloom 5–6 days after heading. The percentage of non-flowering spikelets was 24–36% in BMS10 progenies but only 4.5% in wild type (Fig. 6).
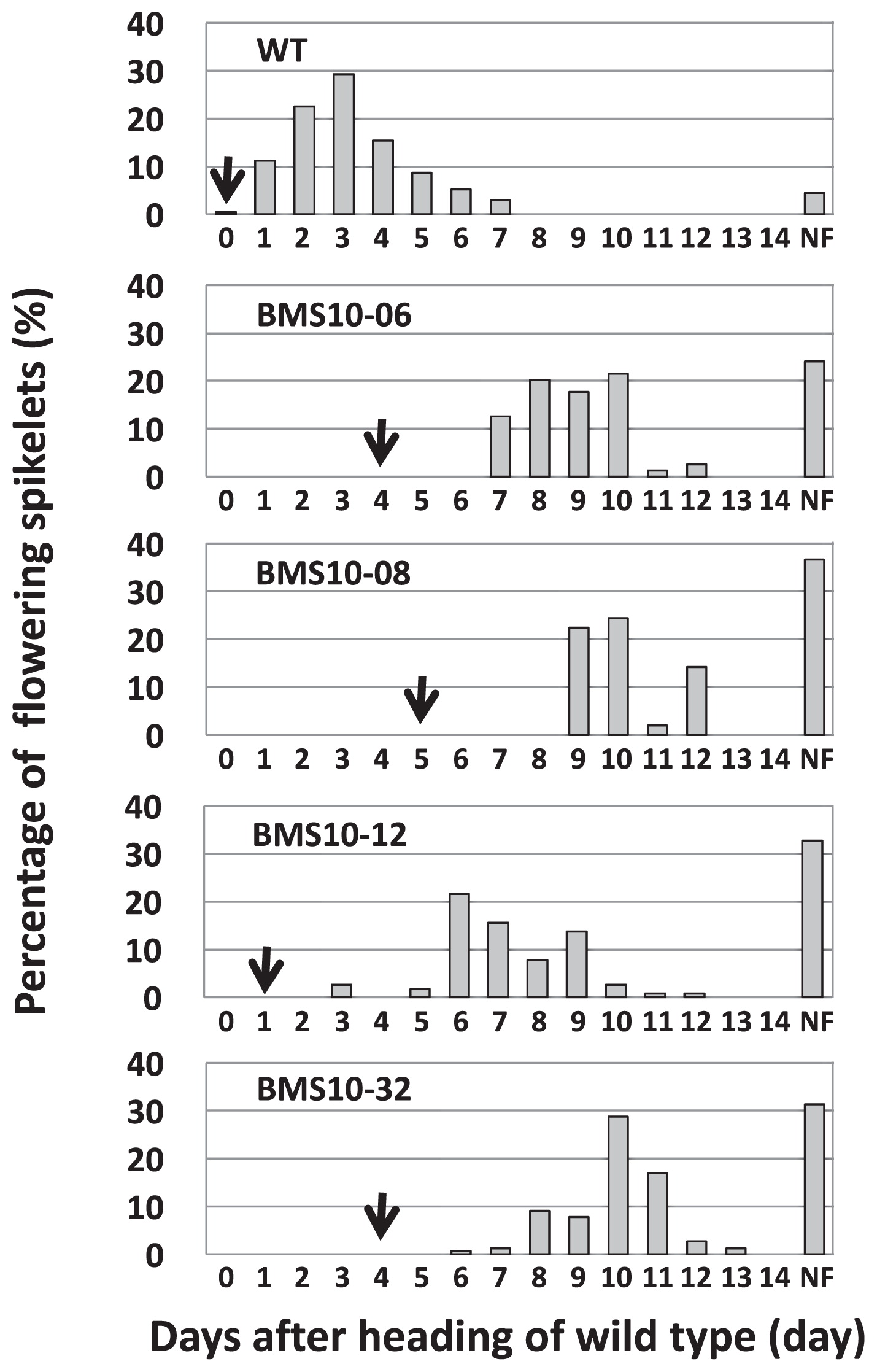
Heading date and percentage of flowering spikelets on each day of wild type and T1 individuals of BMS10. The horizontal axis shows the heading date of wild type as a starting point (0 day). Each arrowhead indicates the heading date of wild type (WT) and BMS10. NF: non-flowering spikelets.
The daily flowering time of BMS progenies and wild type were evaluated (Fig. 7). In wild type plants, 93% of spikelets flowered from 10:00 to 13:00 h. However, 28–60% of spikelets flowered during the same period as BMS individuals. These results indicate that flowering habits, including heading date, flowering date, and daily flowering time of BMS10 progenies tended to be delayed compared to a wild type.
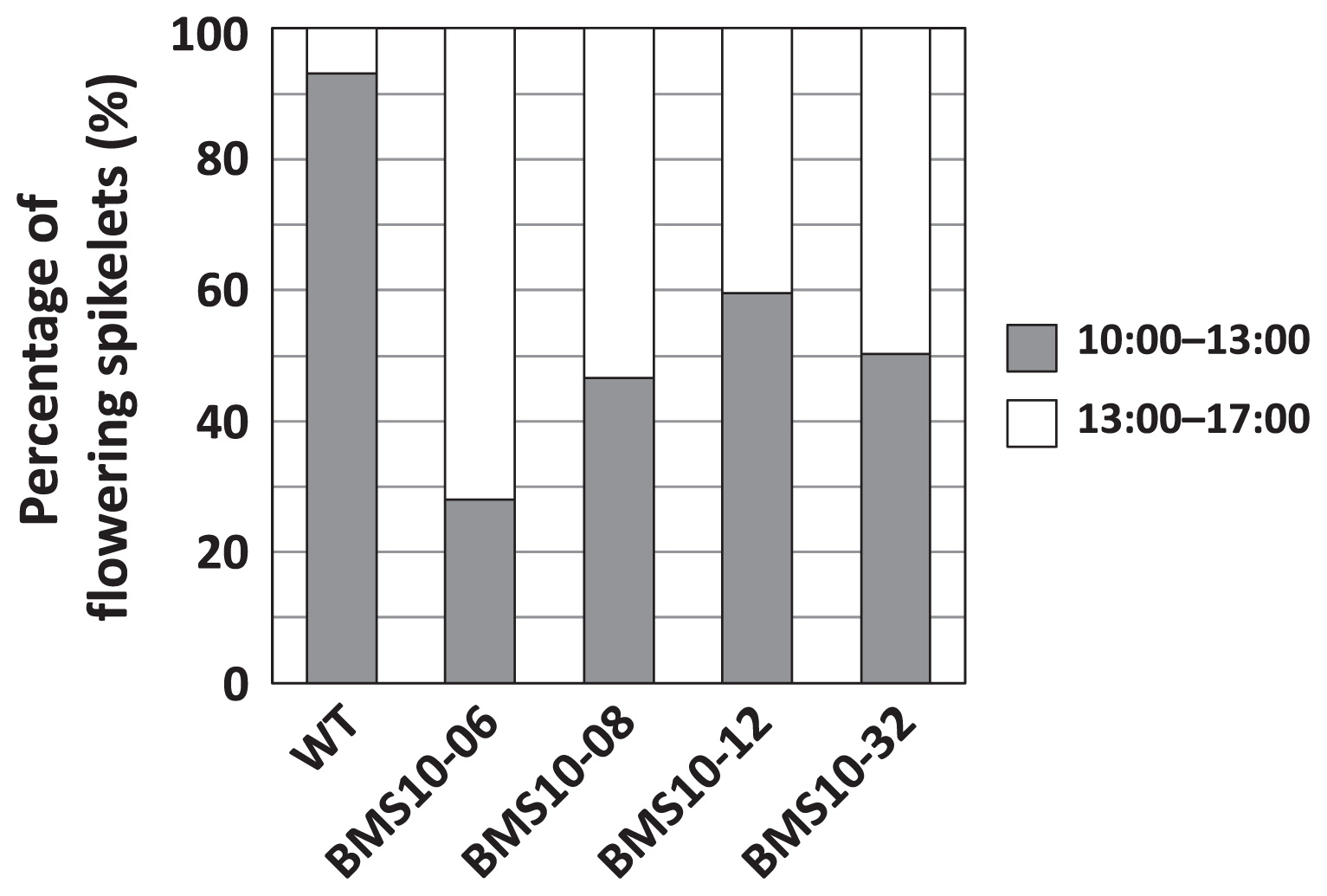
Daily flowering time of wild type (WT) and T1 individuals of BMS10.
Progenies with transgenes of T1 generation in BMS10 and wild type were tied in a bundle and enclosed in a bag. The seed-setting rate of BMS10 progenies scored from 0% to 12.8% (Table 3). The highest seed-setting rate in BMS10-12 was 12.8%. That of BMS10-32 and BMS10-08 was 2.7% and 1.0%, respectively. BMS10-06 did not produce any seeds.
| Cross combination | No. of spikelets | No. of seeds | Seed setting rate (%) |
|---|---|---|---|
| BMS10-06 (self-pollination)a | 65 | 0 | 0.0 |
| BMS10-06 × WTb | 185 | 0 | 0.0 |
| BMS10-08 (self-pollination)a | 57 | 0 | 0.0 |
| BMS10-08 × WTb | 231 | 3 | 1.0 |
| BMS10-12 (self-pollination)a | 39 | 0 | 0.0 |
| BMS10-12 × WTb | 187 | 24 | 12.8 |
| BMS10-32 (self-pollination)a | 56 | 0 | 0.0 |
| BMS10-32 × WTb | 224 | 6 | 2.7 |
WT: Wild type.
Further, the seed-setting rate of BMS10 progenies was examined in another condition. BMS10 progenies and wild type were placed side-by-side and crossed under an open-pollinating condition without cutting the spikelets. Thirty-six BMS10 progenies were used and the seed-setting rate was 0–1.49%.
Finally, the potential seed-setting rate of outcross was explored in BMS10 T1 individuals using the rice cultivar ‘Tachiaoba’ as the pollen parent, because the heading date of ‘Tachiaoba’ and ‘Nipponbare’ were close in short day and high temperature conditions (Tanaka et al. 2016). The seed-setting rate of BMS10 T1 individuals was 34.7–86.4% by the clipping method and enclosed in a bag (Table 4), and these results were far higher than those of other conditions without the clipping method. In addition, a parental test was performed for F1 seedlings by SSR marker (Supplemental Fig. 1).
| Cross combinationa | No. of spikelets | No. of seeds | Seed setting rate (%) |
|---|---|---|---|
| BMS10-101 × Tachiaoba | 66 | 34 | 51.5 |
| BMS10-102 × Tachiaoba | 44 | 38 | 86.4 |
| BMS10-103 × Tachiaoba | 49 | 17 | 34.7 |
| BMS10-104 × Tachiaoba | 89 | 57 | 64.0 |
| BMS10-105 × Tachiaoba | 81 | 50 | 61.7 |
| Total | 329 | 196b | – |
Breeding programs based on repeated outcrosses are desirable for a genomic selection application, even for autogamous crops, such as rice, wheat, and soybean. The final purpose of this study was to contribute a novel breeding system based on repeated outcrosses in rice. In this study, we generated dominant male sterile transgenic rice using barnase and barstar genes. All T0 individuals examined exhibited stable male sterility with normal female fertility (Tables 1–4, Figs. 3, 4). This result indicates that this strategy to produce transgenic dominant male sterility is suitable for rice. Although some T0 individuals showed abnormal or inferior growth, it is thought that some of these might be caused by a leaky expression of the barnase gene in vegetative tissues or somaclonal variation. GUS expression was not observed in the vegetative tissue of rice in a histochemical assay (data not shown). However, male BMS sterility was caused by the weak expression of barnase in the anther (Fig. 2). This observation suggested that barnase with undetectable expression level in the GUS assay affected the negative growth of vegetative organs in BMS. In addition, since there is the possibility that the CaMV35S promoter does not sufficiently work to cancel the influence of barnase in some vegetative rice tissues (Battraw and Hall 1990, Terada and Shimamoto 1990), the expression ratio of barnase and barstar genes in each tissue is important for normal plant growth. However, there is no information about the relationship between plant growth and expression ratio of these genes. We also did not discuss this topic, but focused on the flowering habits and seed-setting rate in normal growth transgenic rice.
Synchronizing heading dates and daily flowering time is important for the efficient production of hybrid seeds by outcross in natural conditions, because the pollen viability of rice is lost within 30 min (Song et al. 2001). Thus, we investigated the flowering habits of the T1 generation in BMS10 and found that flowering habits, including heading date, flowering date, and daily flowering time tended to be delayed compared to that of wild type (Figs. 6, 7). These observations of the BMS10 T1 generation are consistent with other male sterile rice, such as cytoplasmic male sterility and genetic male sterility (Tamaru 1994, Yan and Li 1987). Tamaru (1994) studied the flowering habits of about 29 genetic male sterile rice mutants induced by gamma irradiation or an ethyleneimine treatment, and found that the flowering date and flowering period of these mutants was delayed and that the daily flowering time was extended from early morning to late afternoon.
When BMS10 T1 individuals were crossed with ‘Nipponbare’ and enclosed in a bag without the clipping method, the seed-setting rate was 0–12.8%, and BMS10-12 showed the highest seed-setting rate (Table 3). These results were thought to be due to a different heading date and daily flowering time between BMS10 and wild type. The heading date of BMS10-12 was closest to the wild type; it was 1 day later than that of the wild type and the flowering period of BMS10-12 lines also overlapped with that of the wild type (Fig. 6). Furthermore, about 60% of BMS10-12 spikelets flowered from 10:00 to 13:00 h (Fig. 7). The synchrony of BMS10-12 to wild type might result in higher seed productivity. It can be concluded that flower synchrony is a crucial factor in increasing the seed-setting rate.
When T1 individuals of BMS10 and ‘Nipponbare’ were placed side-by-side and crossed without the clipping method, the seed-setting rate was less < 1.5% (Table 4). This result was lower than that of other reports (Fujimaki et al. 1977). Fujimaki et al. (1977) also examined the seed-setting rate of male sterile mutants and reported that 1.7–8.3% seeds to spikelets were produced. We supposed that the difference in seed-setting rate between ours and their results was caused by the percent synchronization in flowering habits between male sterile rice and the pollen parent, resulting from male sterility. Tamaru (1994) classified several male sterile mutants and investigated the relationship between flowering habits and at what pollen development stage an abnormality occurred in each mutant. The mutants that showed an abnormality at an early stage of pollen development without pollen grains exhibited a delay in heading date and flowering time. Conversely, other mutants that showed an abnormality at the later stage of pollen development with aberrant pollen grains and they exhibited flowering habits similar to wild type. We speculate that sufficient anther and pollen development are important for flowering date and daily flowering time. A histochemical GUS analysis showed that the BoA9 promoter works mainly from an earlier stage to the tetrad stage of pollen development in rice (Fig. 2A, 2B), and this result is similar to the study of the A9 promoter in B. napus (Scott et al. 1991). The BoA9 promoter was used to induce male sterility in BMS, and these BMS did not have pollen grains.
The number of non-flowing spikelets of T1 BMS individuals tended to be higher than that of the wild type (Fig. 6). This difference was observed in a previous study of some male sterile lines (Tamaru 1994). Tamaru (1994) discussed the possibility that increasing non-flowering spikelets does not affect pollen abnormality, but this phenomenon was often observed in each male sterile line. Actually, the T1 generation of BMS showed various percentages of non-flowering spikelets (Fig. 6). Therefore, we could select more practical BMS individuals, i.e., those that showed a small percentage of non-flowering spikelets among all BMS individuals.
To confirm the outcross rate with another rice variety, BMS10 was crossed with ‘Tachiaoba’ by the clipping method to avoid the influence of flowering habits. The seed-setting rate was 34.7–86.4% (Table 4) and a total 94 progenies from four individuals (BMS10-101–BMS10-104) were confirmed (Supplemental Fig. 1). This result demonstrated that any negative influence of the barnase protein was not observed in BMS10. This result is also an important finding for the development of a tool to obtain hybrid seeds without emasculation such as hot water emasculation. Hot water emasculation consists of four steps as follows: (1) hot water emasculation, (2) removing the spikelets that have already flowered to avoid seed setting by self-pollination, (3) clipping the unflowered spikelets and (4) pollination. If we can use male sterile rice as a seed parent, the above-mentioned steps (1) and (2) are not needed. In addition, the use of male sterile rice as a seed parent can avoid the damage of the pistil tissue resulting from the hot water emasculation. Thus, transgenic male sterile rice has important advantages for crossing compare to hot water emasculation.
To improve flowering habits and seed-setting rates, we propose that another anther-specific promoter is used for the expression of barnase. For example, male sterile rice will exhibit similar flowering habits to wild type when the promoter induces genes at a later developmental stage during anther development. Another approach to resolve the low seed-setting rate is the introduction of a stigma exsertion phenotype combined with transgenic male sterility. Exserted stigma exposed outside of the spikelets maintained activities for 6 days after the spikelets closed in rice (Kato and Namai 1987, Yan et al. 2009). Although stigma exsertion cannot contribute to the synchronization of flowering habits, it seems to be effective for increasing pollination and the seed-setting rate.
Tanaka (2010) proposed the utilization of positive selection, e.g., a herbicide-tolerance gene driven by a constitutive promoter and negative selection, e.g., a lethal gene driven by an inducible promoter for the selection of fertile or sterile progeny. We consider that this proposal should improve the outcross system if we can introduce those expression cassettes with the male sterile induction cassette into one locus of the genome. We believe that the novel breeding tool described here would contribute to repetitive outcross and facilitate efficient and practical genomic selection in autogamous crop populations.
We thank Ms. I. Kawaguchi, Ms. C. Ito, Ms. R. Masuda, Ms. A. Sugai, Ms. K. Sasaki, and Ms. E. Mitsuhara for their technical help. This work was supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation GMO-1001).