2021 Volume 71 Issue 2 Pages 155-166
2021 Volume 71 Issue 2 Pages 155-166
Seed dormancy, a major factor regulating pre-harvest sprouting, can severely hinder wheat cultivation. Reduced Seed Dormancy 32 (RSD32), a wheat (Triticum aestivum L.) mutant with reduced seed dormancy, is derived from the pre-harvest sprouting tolerant cultivar, ‘Norin61’. RSD32 is regulated by a single recessive gene and mutant phenotype expressed in a seed-specific manner. Gene expressions in embryos of ‘Norin61’ and RSD32 were compared using RNA sequencing (RNA-seq) analysis at different developmental stages of 20, 30, and 40 days after pollination (DAP). Numbers of up-regulated genes in RSD32 are equivalent in all developmental stages. However, down-regulated genes in RSD32 are more numerous on DAP20 and DAP30 than on DAP40. In central components affecting the circadian clock, homologues to the morning-expressed genes are expressed at lower levels in RSD32. However, higher expressions of homologues acting as evening-expressed genes are observed in RSD32. Homologues of Ca2+ signaling pathway related genes are specifically expressed on DAP20 in ‘Norin61’. Lower expression is shown in RSD32. These results suggest that RSD32 mutation expresses on DAP20 and earlier seed developmental stages and suggest that circadian clock regulation and Ca2+ signaling pathway are involved in the regulation of wheat seed dormancy.
Wheat, similarly to rice and maize, is a major crop worldwide that is crucially important for world food supplies. Pre-harvest sprouting, which is triggered by continuous rainfall during seed development, occurs as seed germination on mother plants. This germination decreases seed quality and causes extensive economic damage to cultivation efforts. Seed dormancy is a major regulating factor affecting pre-harvest sprouting. Seed dormancy, which inhibits seed germination under favorable conditions, particularly of temperature and moisture, occurs after completion of seed maturation (Gubler et al. 2005). Therefore, enhancing seed dormancy is an important breeding objective for avoiding pre-harvest sprouting damage.
Seed dormancy begins and develops during seed maturation. The embryo axis and scutellum differentiate and grow extensively at an early developmental stage (5–15 days after pollination: DAP5–DAP15) in wheat (Noda et al. 1994). Between DAP15 and DAP20, the embryo dry weight and embryo axis length increase rapidly. The length reaches its maximum at DAP30. The embryo appears to be fully differentiated at DAP30. The endosperm fresh and dry weight reach maximum values at DAP30. Seed development is completed at the middle developmental stage (DAP15–DAP30). At the late developmental stage (DAP30–DAP50), seed moisture contents decrease. Endosperms reach the hard dough stage. Seeds desiccate and change color from yellow to brown. Seed dormancy develops during seed desiccation in the late developmental stage.
A phytohormone, abscisic acid (ABA), plays an important role in controlling seed dormancy. In Arabidopsis, many seed-dormancy mutants have been isolated. Results of earlier studies have shown that ABA biosynthesis, catabolism, and sensitivity are involved in regulating seed dormancy (Finkelstein et al. 2002, Himmelbach et al. 2003, Kushiro et al. 2004, Nambara and Marion-Poll 2003, Okamoto et al. 2006, Saito et al. 2004, Seo et al. 2006). DELAY OF GERMINATION 1 (DOG1) has been identified as a quantitative trait locus (QTL) controlling the natural variation of seed dormancy in Arabidopsis (Bentsink et al. 2006). In fact, DOG1 interacts with ABA signaling pathway through type 2C protein phosphatases ABA-HYPERSENSITIVE GERMINATION 1 (AHG1) and AHG2 (Née et al. 2017, Nishimura et al. 2018). Seed maturation regulators LEAFY COTYLEDON 1 (LEC1), LEC2, FUSCA3 (FUS3) and ABA INSENSITIVE 3 (ABI3) are also involved in the regulation of seed dormancy (Giraudat et al. 1992, Kagaya et al. 2005, Kroj et al. 2003, Lotan et al. 1998, Luerßen et al. 1998, Stone et al. 2001, To et al. 2006). These genes express at the early to late developmental stages of seed in Arabidopsis. In monocot species, MOTHER OF FT AND TFL1 (MFT) and MAP KINASE KINASE in wheat (Nakamura et al. 2011, Torada et al. 2016), SDR4 in rice (Sugimoto et al. 2010), and ALANINE AMINOTRANSFERASE (AlaAT) in barley (Sato et al. 2016) have been identified as QTLs regulating seed dormancy. Rikiishi et al. (2010) reported that TaABF1 related with ABA signaling pathway regulates intervarietal variation of seed dormancy in wheat cultivars. Several genes regulating seed dormancy have also been identified in wheat. These genes express at the late seed developmental stage. However, wheat genes homologous to DOG1 and seed maturation regulators are expressed at the early to middle seed development stage (Rikiishi and Maekawa 2014). The appropriate time for expression differs depending on the regulator gene. These results indicate that different regulatory systems for seed dormancy regulation function at each developmental stage.
Reports of the literature describe that ABA signaling is connected with and integrated with other signaling pathways. Calcium ion acts as a second messenger. In fact, calcium signals in plants are involved in several stress responses to cold, drought, salinity and light (Dodd et al. 2010, Zhang et al. 2014). The Ca2+ signaling pathway is initiated with the acceptance of Ca2+ signals by sensor proteins. Plant Ca2+ sensors belong to three families (Edel and Kudla 2015, Zhu et al. 2015). Calmodulins (CaMs) and CaM-like proteins (CMLs) are grouped in one family. The second family includes calcineurin B-like proteins (CBLs) that specifically activate CBL-interacting protein kinases (CIPKs). The third family includes Ca2+-dependent protein kinases (CDPKs), which have a kinase domain and a Ca2+ sensor domain. Sensor proteins accepting Ca2+ signals are decoded to downstream responses. Because Ca2+ sensor proteins affect ABA sensitivity, Ca2+ signaling cooperatively regulates the response to stresses with the ABA signaling pathway (Chen et al. 2017, Edel and Kudla 2016, Jiang et al. 2013, Midhat et al. 2018, Wang et al. 2018, Zhao et al. 2011). Several reports have described functions of circadian-clock-related genes on ABA sensitivity and dormancy release (Adams et al. 2018, Footitt et al. 2017, Lee et al. 2016, Penfield and Hall 2009, Seung et al. 2012). The circadian clock regulates the gene expressions and physiological responses corresponding to a daily cycle of light and darkness. In Arabidopsis, LATE ELONGATED HYPOCOTYL (LHY), CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), TIMING OF CAB EXPRESSION1/PSEUDO-RESPONSE REGULATOR (TOC1/PRR), EARLY FLOWERING (ELF) and LUX ARRHYTHMO (LUX) are involved in the central cores of the circadian clock (Seung et al. 2012). These genes encode transcription factors or proteins forming complexes with transcription factor and construct a complex system with feedback loop regulation. Circadian rhythms oscillate based on interactions between morning-expressed LHY and CCA1 and evening-expressed TOC1. Circadian clock components interact with various transcription factors such as NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED 1 (LNK1), FAR-RED IMPAIRED RESPONSE 1 (FAR1), PHYTOCHROME INTERACTING FACTORs (PIFs), REVEILLEs (RVEs), and CONSTANS-LIKE (Gray et al. 2017, Ledger et al. 2001, Nusinow et al. 2011, Ritter et al. 2017, Xing et al. 2015). Furthermore, circadian clock genes regulate cytosolic Ca2+ influx and the signaling pathways of ABA and Ca2+, suggesting that regulatory network integration is necessary for circadian-clock-related fundamental processes in plant growth and development (Dodd et al. 2005, 2007, Martí Ruiz et al. 2018, Xu et al. 2007).
Rikiishi and Maekawa (2010) produced a wheat (Triticum aestivum L.) mutant with reduced seed dormancy from ‘Norin61’, a pre-harvest sprouting tolerant cultivar. Reduced Seed Dormancy 32 (RSD32) was found to be a seed-specific and single-recessive mutation (Kobayashi et al. 2006, Rikiishi and Maekawa 2010). Expressions of several transcription factors involved in the regulation of wheat seed dormancy, such as TaDOG1 and TaABF1, were decreased in embryos of RSD32, suggesting RSD32 as an important factor for the regulation of seed dormancy in wheat.
In this study, gene expressions in embryos of ‘Norin61’ and RSD32 were analyzed using RNA-seq for investigating the regulatory networks of wheat seed dormancy associated with RSD32. Expression profiles were compared at three developmental stages: DAP20, DAP30, and DAP40. Results demonstrate that RSD32 mutation exhibits superior inhibitory effects on gene expression in embryos on DAP20 and DAP30. In embryos of RSD32, homologous genes of circadian clock and Ca2+ signaling pathway related genes were expressed differently compared to ‘Norin61’. RSD32 is a regulatory factor for wheat seed dormancy expressed at the middle developmental stage. Reduced seed dormancy in RSD32 might result from aberrant signals of the circadian clock and Ca2+.
This study used a pre-harvest sprouting tolerant wheat cultivar, ‘Norin61’, and a mutant RSD32 with reduced seed dormancy selected from M4 population. They were derived from mutagenized ‘Norin61’ seeds using NaN3 treatment (Rikiishi and Maekawa 2010). Seeds were sown in plastic trays for 4 weeks: later, 20 seedlings were transplanted to the field in each line with 20 cm between plants and 90 cm between rows. Plants were grown under a plastic roof to avoid rainfall. Spikes were tagged at anthesis. Seeds were harvested every 10 days from 10 days after pollination (DAP10) to DAP70 and were used in germination tests and RNA-seq analysis. To minimize variation, seeds were collected only from primary and secondary florets of the center spikelets.
Germination testTen whole seeds were placed on filter paper in a Petri dish containing 6 ml of distilled water. Seeds were cut transversely into halves. Then ten half seeds with involved embryos were placed in a Petri dish containing 6 ml of distilled water with or without (±) 10 μM of ABA (Sigma Chemical Co.). The Petri dishes were then incubated in the dark at 20°C. All germination tests used three replications. Germinated seeds were counted daily for 14 days. A weighted germination index (GI) was calculated to give maximum weight to seeds that germinated first and to give less weight to those that germinated subsequently, as described by Walker-Simmons and Sesing (1990). The GI values were converted into arcsine-transformed values. They were used for statistical analyses.
RNA isolation, library preparation and sequencingEmbryos of ‘Norin61’ and RSD32 were collected from 10:00 to 12:00 on the same day. Three embryos from a single panicle were used for total RNA isolation. On DAP20, DAP30, and DAP40 panicles were harvested from three individual plants. Each plant was treated as biological replication. Total RNA was isolated using a commercial kit (FastRNA Pro Green; Qbiogene Inc.). Isolated RNAs were purified (RNA Clean-up Kit; TaKaRa Bio Inc., Tokyo, Japan). All kits were used according to the respective manufacturers’ protocols. The concentrations of total RNA samples were quantified using a spectrophotometer (Nano Drop ND-1000; Thermo Fisher Scientific Inc., Waltham, MA, USA). The total RNA sample quality was also verified (Agilent 2100 Bioanalyzer; Agilent Technologies Inc., Santa Clara, CA, USA). The 18 RNA samples (2 lines × 3 stages × 3 biological replications) were sequenced. Library construction and sequencing for the Illumina HiSeq 2500 were provided as a custom service of Eurofins Genomics K.K. (Tokyo, Japan). After the polyA fraction (mRNA) was isolated from total RNA, it was fragmented. Then double-stranded (ds) cDNA was reverse-transcribed from the fragmented mRNA. The ds cDNA fragments were processed for adaptor ligation, size selection (for 200 bp inserts), and amplification to generate strand-specific cDNA libraries. Prepared libraries were subjected to paired-end 2 × 125 bp sequencing on the HiSeq 2500 platform with v4 chemistry.
Bioinformatics analysisWe analyzed RNA-seq read data using RNA analysis tools in Galaxy/NAAC (https://galaxy.dna.affrc.go.jp/). Raw reads were obtained in Fastq format and were assessed for quality using FastQC. Terminal low-quality bases and adaptor sequences were trimmed off (Trimmomatic; Usadel Lab, Aachen, Germany). Clean reads were aligned against wheat survey sequence v3.0 obtained from the International Wheat Genome Sequencing Consortium (IWGSC) using TopHat2 with default parameters (Kim et al. 2013a, Trapnell et al. 2012). Cufflinks was used to assemble mapped reads. The resulting transcripts were used to quantify the expression of each gene in fragments per kilobase of transcript per million mapped reads (FPKM) units. Cuffdiff was subsequently used to compile a list of differentially expressed genes (DEGs) with fold change ≥3 and P-value ≤0.01. BLASTX was used to align genes against the National Center of Biotechnology Information (NCBI) database (https://blast.ncbi.nlm.nih.gov/Blast.cgi), the Rice Annotation Project Database (https://rapdb.dna.affrc.go.jp/tools/blast), Wheat Genetic Resources Database (https://shigen.nig.ac.jp/wheat/komugi/blast/blast.jsp), and Barley BioResources Database (http://earth.nig.ac.jp/~dclust/cgi-bin/barley_pub/blast_search.html). The e-value cutoff was set at 1e–5. Gene names were assigned to each gene based on the top Blastx hit with the highest score. RStudio (https://rstudio.com/) was used for comparing DEGs in three developmental stages.
Anthesis was observed from 27 April through 16 May in RSD32, similarly to wild type, ‘Norin61’ (Supplemental Fig. 1). Morphological traits of panicles and seeds were not different between ‘Norin61’ and RSD32 (Supplemental Fig. 2). Although heading time and seed development affect the degree of seed dormancy, no disorders of these traits were observed in RSD32.
‘Norin61’ showed low germination indices (GIs) of whole seeds obtained at DAP50 and earlier stages. Strong seed dormancy was observed (Fig. 1). The GIs of half seeds, which were released from dormancy, were significantly lower with ABA treatment until DAP50. ‘Norin61’ showed sensitivity to ABA on seed germination. Seed dormancy and ABA sensitivity were lost after DAP60 in ‘Norin61’. However, RSD32 showed significantly higher GIs of whole seeds on DAP40 (50.0) and on DAP50 (90.5) than those in ‘Norin61’, although similar GIs of whole seeds were detected on DAP10–DAP30. Results show that RSD32 revealed the reduced seed dormancy phenotype at late developmental stages. The GIs of half seeds were 86.9 and 95.2, respectively, on DAP40 in ‘Norin61’ and RSD32 and remained at higher levels at later stages. Inhibitory effects of ABA on germination were reduced in RSD32 on DAP20, DAP30, and DAP50. These results indicate that RSD32 showed similar degrees of seed dormancy to those of ‘Norin61’ on DAP20 and DAP30. However, seed dormancy was found to be markedly lower on DAP40–DAP50 in RSD32. Reduced ABA sensitivity was also detected on DAP50 in RSD32.

Germination index (GI) whole seeds in water and half seeds in water with and without 10 μM ABA in ‘Norin61’ and RSD32 at different developmental stages. Error bars represent SE.
Reduction of seed dormancy was detected on DAP40 and DAP50 in RSD32. Gene expression was compared using RNA-seq analysis at the middle to late developmental stages: DAP20, DAP30, and DAP40. Numbers of DEGs that were down-regulated in RSD32 were 209, 228, and 49, respectively, on DAP20, DAP30, and DAP40 (Fig. 2). Down-regulated genes in RSD32 were detected more on DAP20 and DAP30 than on DAP40. Numbers of DEGs that were up-regulated in RSD32 were similar at all developmental stages. RSD32 mutation preferentially inhibited gene expression. Marked effects were observed at earlier developmental stages than on DAP40 when seed dormancy reduction started.
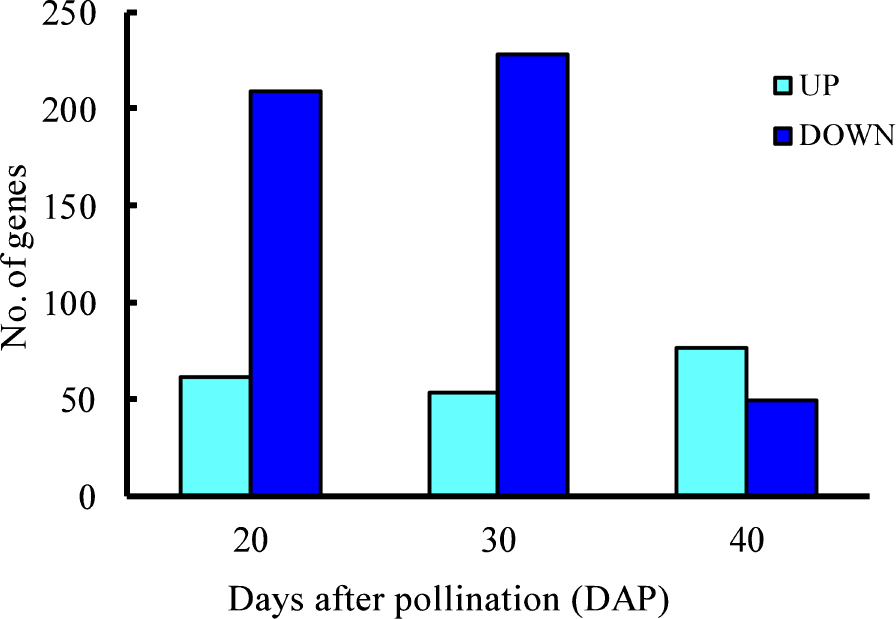
Numbers of differentially expressed genes (DEGs) between embryos of ‘Norin61’ and RSD32 at different developmental stages. UP and DOWN respectively denote up-regulated and down-regulated genes in RSD32.
Comparison of up-regulated genes in RSD32 showed less overlap among developmental stages. Actually, most up-regulated genes were expressed specifically at each developmental stage (Fig. 3A). Examination of down-regulated genes in RSD32 among developmental stages revealed that 146 and 164 DEGs, respectively, showed specific expression on DAP20 and DAP30, and that 62 DEGs were down-regulated at DAP20 and DAP30 (Fig. 3B). Most down-regulated genes on DAP40 showed stage-specific expression. No overlap with other developmental stages was observed.

Venn diagram highlighting the number of differentially expressed genes in the three developmental stages. UP and DOWN respectively denote up-regulated and down-regulated genes in RSD32.
At DAP20, down-regulated genes in RSD32 revealed several functions such as gene expression, protein metabolism, oxidation–reduction process, response to stimuli, circadian rhythm, and signal transduction (Tables 1, 2). These genes involved several homologous genes related to the calcium signaling pathway, such as CALCIUM-BINDING PROTEIN, CALMODULIN-BINDING PROTEIN, EF-HAND Ca2+-BINDING PROTEIN, CALMODULIN-BINDING RECEPTOR-LIKE CYTOPLASMIC KINASE, CALCIUM-DEPENDENT PROTEIN KINASE, and CALCIUM-TRANSPORTING ATPASE. Furthermore, down-regulated genes involved circadian-clock-related genes. Genes homologous to CCA1, LHY, LNK1, and RVE6-LIKE were expressed lower in RSD32 than in ‘Norin61’. Down-regulated genes were classified into Ca2+ signal transduction (16 genes) and circadian-clock regulation (14 genes) more than the responses to hormones (9 genes) and light (6 genes). Although many genes of heat shock protein family were observed in down-regulated genes on DAP20 (44 genes) and DAP30 (73 genes), their functions remained unknown. In fact, Ca2+ signal transduction and circadian-clock regulation were the leading functional groups in down-regulated genes on DAP20. Circadian-clock-related genes were also identified as up-regulated genes in RSD32. Genes homologous to FAR1-RELATED SEQUENCE 12-LIKE and CONSTANS-LIKE 9 were found to be expressed higher in RSD32 than in ‘Norin61’ on DAP20 (Table 3). A homologous gene to TOC1 was also expressed 2.4 times higher in RSD32. These results indicate that unusual calcium signaling and circadian-clock regulation occurred in RSD32.
| Putative function | FPKM* | Fold change | |
|---|---|---|---|
| ‘Norin61’ | RSD32 | ||
| Cellular component biogenesis | |||
| WAT1-related protein/auxin-induced protein 5NG4 | 80.4 | 0 | – |
| Cellular metabolic process | |||
| haloacid dehalogenase-like hydrolase domain-containing protein 3 | 2235.6 | 744.7 | 3.0 |
| pheophorbide a oxygenase | 107.9 | 29.5 | 3.7 |
| photosystem II reaction center PSB28 | 62.7 | 0 | – |
| soluble inorganic pyrophosphatase | 312.8 | 40.6 | 7.7 |
| Cellular process | |||
| CDGSH iron-sulfur domain-containing protein NEET | 160.2 | 45.6 | 3.5 |
| CytHSP70 | 63.1 | 5.1 | 12.3 |
| hypoxia-induced gene domain 5 | 773.8 | 231.1 | 3.3 |
| Circadian rhythm | |||
| LNK1 | 766.5 | 198.7 | 3.9 |
| LNK2 | 125.7 | 39.9 | 3.2 |
| LNK4-like | 554.4 | 129.2 | 4.3 |
| Developmental process | |||
| Senescence associated gene 20 (SAG20) | 231.0 | 45.5 | 5.1 |
| Gene expression | |||
| antagonist of like heterochromatin protein 1-like (ALP1) | 53.9 | 15.6 | 3.4 |
| B-box zinc finger protein 25-like | 356.3 | 108.8 | 3.3 |
| calmodulin-binding protein 60 D-like | 213.9 | 46.0 | 4.6 |
| lariat debranching enzyme | 762.0 | 154.6 | 4.9 |
| light-inducible protein CPRF2 | 436.7 | 127.3 | 3.4 |
| multiprotein bridging factor 1 (MBF1) | 530.2 | 81.4 | 6.5 |
| NAC domain-containing protein 74 | 574.9 | 127.1 | 4.5 |
| pentatricopeptide repeat-containing protein | 66.3 | 16.5 | 4.0 |
| PsbB mRNA maturation factor Mbb1 | 721.4 | 169.4 | 4.3 |
| ribosomal protein S8 | 712.9 | 224.4 | 3.2 |
| RNA-binding motif protein | 278.0 | 47.2 | 5.9 |
| WRKY11 transcription factor | 97.1 | 0 | – |
| Localization | |||
| calcium-transporting ATPase 1 | 199.7 | 55.3 | 3.6 |
| glucose-6-phosphate/phosphate-tranlocator-like | 192.4 | 36.7 | 5.2 |
| P-type ATPase | 215.6 | 68.5 | 3.1 |
| Molecular function regulator | |||
| kelch repeat-containing protein-like | 191.3 | 38.6 | 5.0 |
| Organic substance metabolic process | |||
| phosphoglycerate mutase gpmB | 112.8 | 36.1 | 3.1 |
| Oxidation–reduction process | |||
| 3-beta hydroxysteroid dehydrogenase/isomerase | 253.2 | 70.3 | 3.6 |
| Aldehyde dehydrogenase (ALDH) | 186.8 | 57.7 | 3.2 |
| cytochrome P450 | 70.3 | 21.7 | 3.2 |
| L-ascorbate oxidase | 86.5 | 20.0 | 4.3 |
| NAD(P)-binding Rossmann-fold protein | 149.6 | 48.5 | 3.1 |
| NADH dehydrogenase | 855.1 | 269.2 | 3.2 |
| nitrate reductase [NAD(P)H] | 313.3 | 44.9 | 7.0 |
| omega-3 fatty acid desaturase | 228.9 | 71.7 | 3.2 |
| premnaspirodiene oxygenase-like | 162.0 | 0 | – |
| respiratory burst oxidase B-like | 372.0 | 62.3 | 6.0 |
| retinal dehydrogenase | 72.8 | 0 | – |
| Protein metabolic process | |||
| ADP-ribosylation factor | 74.2 | 14.6 | 5.1 |
| ankyrin repeat-containing protein NPR4 | 305.4 | 85.5 | 3.6 |
| Aspartic proteinase Asp1 | 303.3 | 61.3 | 4.9 |
| calcium-dependent protein kinase | 1010.3 | 184.9 | 5.5 |
| E3 ubiquitin-protein ligase | 201.2 | 65.0 | 3.1 |
| Leaf rust 10 disease-resistance locus receptor-like protein kinase | 98.4 | 0 | – |
| prolyl 4-hydroxylase 6 precursor | 282.7 | 54.4 | 5.2 |
| receptor-like protein kinase | 165.4 | 54.5 | 3.0 |
| RHOMBOID-like protein 2 | 537.6 | 132.3 | 4.1 |
| wall-associated receptor kinase 2-like (WAK2-like) | 159.8 | 44.7 | 3.6 |
| Response to stimulus | |||
| 16.9a kDa heat-shock protein | 501.7 | 152.5 | 3.3 |
| ATA15 | 630.6 | 201.9 | 3.1 |
| Early responsive to dehydration 15-like | 1738.5 | 461.4 | 3.8 |
| heat shock cognate 70 kDa protein 2-like | 443.6 | 115.3 | 3.8 |
| heat-responsive transcription factor | 284.0 | 90.1 | 3.2 |
| small EDRK-rich factor 2-like (SERF2) | 1256.2 | 376.9 | 3.3 |
| EXORDIUM-like | 184.8 | 19.9 | 9.3 |
| hemoglobin Hb2 | 298.8 | 78.7 | 3.8 |
| ethylene-responsive transcription factor ERF071-like | 215.8 | 46.9 | 4.6 |
| indole-3-acetic acid-amido synthetase (GH3.3) | 367.6 | 110.8 | 3.3 |
| RVE6-like | 241.9 | 48.5 | 5.0 |
| Secondary metabolic process | |||
| phenylalanine ammonia-lyase-like | 104.1 | 11.0 | 9.5 |
| Signal transduction | |||
| calcium-binding protein | 93.6 | 0 | – |
| calmodulin-binding receptor-like cytoplasmic kinase 3 | 114.2 | 33.2 | 3.4 |
| calmodulin-related protein | 581.5 | 133.8 | 4.3 |
| CBL-interacting protein kinase 31 | 358.5 | 103.7 | 3.5 |
| EF-hand Ca2+-binding protein CCD1 | 334.6 | 81.5 | 4.1 |
| mitogen-activated protein kinase | 1358.4 | 387.6 | 3.5 |
| SOUL heme-binding domain containing protein | 416.0 | 59.6 | 7.0 |
| Others | |||
| retrotransposon protein | 54.5 | 0 | – |
| serine-rich protein | 179.1 | 38.2 | 4.7 |
* FPKM denotes Fragments Per Kilobase of transcript per Million mapped reads.
| Putative function | FPKM* | Fold change | |
|---|---|---|---|
| ‘Norin61’ | RSD32 | ||
| Cellular component biogenesis | |||
| 16.9 kDa class I heat shock protein 1-like | 3522.9 | 815.5 | 4.3 |
| 17.5 kDa class II heat shock protein | 990.7 | 168.8 | 5.9 |
| 17.5 kDa heat-shock protein | 312.7 | 57.3 | 5.5 |
| 17.9 kDa class I heat shock protein | 727.0 | 91.4 | 8.0 |
| 18.6 kDa class III heat shock protein-like | 2564.1 | 693.4 | 3.7 |
| 23.2 kDa heat shock protein-like | 189.4 | 31.3 | 6.0 |
| heat shock protein 16.9 | 1006.8 | 61.4 | 16.4 |
| small heat shock protein 17.3 kDa | 7957.7 | 1891.0 | 4.2 |
| small heat shock protein Hsp23.5 | 1918.5 | 283.3 | 6.8 |
| Cellular process | |||
| 70 kDa peptidyl-prolyl isomerase | 701.2 | 176.5 | 4.0 |
| BAG family molecular chaperone regulator 6 | 99.8 | 18.5 | 5.4 |
| peptidyl-prolyl cis-trans isomerase | 1101.5 | 288.2 | 3.8 |
| regulator of chromosome condensation (RCC1) | 376.7 | 81.8 | 4.6 |
| Circadian rhythm | |||
| CCA1 | 1407.7 | 313.7 | 4.5 |
| LHY | 139.5 | 27.2 | 5.1 |
| Gene expression | |||
| lariat debranching enzyme | 2892.9 | 763.9 | 3.8 |
| zinc finger MYM-type protein 1-like | 283.4 | 62.3 | 4.5 |
| Oxidation-reduction process | |||
| early nodulin-93-like | 421.7 | 111.5 | 3.8 |
| Protein metabolic process | |||
| receptor-like protein kinase | 158.9 | 0 | – |
| Response to stimulus | |||
| 14.5 kDa heat-shock protein | 1341.7 | 387.5 | 3.5 |
| 23.6 kDa heat shock protein | 1410.9 | 214.1 | 6.6 |
| heat shock cognate 70 kDa protein | 93.9 | 7.4 | 12.7 |
| heat shock protein 90 | 1903.2 | 407.3 | 4.7 |
| ultraviolet-B receptor UVR8 | 266.0 | 59.7 | 4.5 |
| universal stress protein PHOS32 | 739.4 | 212.8 | 3.5 |
| RVE6-like | 284.6 | 60.4 | 4.7 |
| Secondary metabolic process | |||
| 4-coumarate--CoA ligase 3 | 138.8 | 22.5 | 6.2 |
* FPKMs represent values obtained on DAP20.
| Putative function | FPKM | Fold change | |
|---|---|---|---|
| ‘Norin61’ | RSD32 | ||
| Cellular component organization | |||
| vasodilator-stimulated phosphoprotein-like | 25.9 | 94.1 | 3.6 |
| Circadian rhythm | |||
| CONSTANS-LIKE 9 | 55.1 | 162.8 | 3.0 |
| Developmental process | |||
| myb-related protein Zm38-like | 50.5 | 153.8 | 3.0 |
| Gene expression | |||
| AT hook motif containing protein | 0 | 49.7 | – |
| dehydration-responsive element-binding protein 2C-like | 561.8 | 1696.4 | 3.0 |
| Eukaryotic translation initiation factor 2 subunit 3 | 0 | 414.3 | – |
| heat shock factor C1b | 52.7 | 279.4 | 5.3 |
| serine/threonine-protein kinase | 0 | 177.8 | – |
| suppressor protein SRP40-like | 50.6 | 224.3 | 4.4 |
| transcription initiation factor TFIID subunit 4-like | 140.2 | 438.3 | 3.1 |
| zinc finger MYM-type protein 1 | 57.8 | 204.7 | 3.5 |
| Multi-organism process | |||
| hyphally regulated protein-like | 0 | 63.6 | – |
| Organic substance metabolic process | |||
| 3-ketoacyl-CoA synthase 12 | 115.4 | 352.0 | 3.1 |
| glyoxalase family like protein | 424.3 | 1473.3 | 3.5 |
| thiamine thiazole synthase 2 | 234.6 | 750.6 | 3.2 |
| Oxidation–reduction process | |||
| cytochrome P450 71C1 | 292.7 | 1483.4 | 5.1 |
| divinyl chlorophyllide a 8-vinyl-reductase | 0 | 65.9 | – |
| Protein metabolic process | |||
| glutathione S-transferase GSTU6 | 738.4 | 2230.3 | 3.0 |
| Histone acetyltransferase HAC12 | 16.1 | 59.9 | 3.7 |
| TNP2-like protein | 295.6 | 1157.1 | 3.9 |
| WD repeat domain phosphoinositide-interacting protein 3 | 34.8 | 140.4 | 4.0 |
| Response to stimulus | |||
| root peroxidase | 35.7 | 198.2 | 5.6 |
| serine proteinase inhibitor-like allergen | 118.3 | 614.7 | 5.2 |
| FAR1-related sequence12-like | 37.0 | 121.2 | 3.3 |
| dormancy-associated protein 1-like/auxin-repressed protein-like protein ARP1 | 2425.0 | 7415.5 | 3.1 |
| Dehydrin DHN3 | 565.1 | 1840.4 | 3.3 |
| Universal stress protein A-like | 125.5 | 382.8 | 3.0 |
| Signal transduction | |||
| microtubule-associated serine/threonine-protein kinase 4-like | 35.1 | 106.2 | 3.0 |
| Others | |||
| transposon protein | 0 | 63.2 | – |
Down-regulated genes found on DAP30 had several functions. Genes homologous to PIF1-LIKE CCA1 and LHY were down-regulated in RSD32 (Table 2, Supplemental Table 1). However, genes homologous to PHYTOCLOCK1 and LUX-B showed higher expression in RSD32 on DAP30 (Table 4). Genes homologous to CONSTANS-LIKE 9 and FAR1-RELATED SEQUENCE 5-LIKE were expressed, respectively, 2.7 times and 2.5 times higher in RSD32. Expressions of genes homologous to CCA1 and LHY were inhibited in RSD32 at both DAP20 and DAP30. Down-regulated genes in RSD32 at both of DAP20 and DAP30 were enriched to HEAT SHOCK PROTEINs (Table 2). Although some DEGs were identified as gene expression, oxidation–reduction-process, and protein-metabolic-process related genes on DAP40, circadian clock related genes were not found in DEGs on DAP40 (Supplemental Tables 2, 3).
| Putative function | FPKM | Fold change | |
|---|---|---|---|
| ‘Norin61’ | RSD32 | ||
| Cellular component biogenesis | |||
| WAT1-related protein | 0 | 34.7 | – |
| extracellular glycosidase CRH11-like | 0 | 391.6 | – |
| Cellular metabolic process | |||
| lipid phosphate phosphatase 3 | 18.8 | 80.0 | 4.3 |
| Circadian rhythm | |||
| LUX-B | 68.5 | 246.9 | 3.6 |
| PHYTOCLOCK1 | 161.1 | 536.4 | 3.3 |
| Developmental process | |||
| myosin-14-like | 0 | 110.3 | – |
| Gene expression | |||
| B3 domain-containing protein | 0 | 218.7 | – |
| trihelix transcription factor GTL1-like | 0 | 369.0 | – |
| zinc finger protein 410 | 0 | 8707.7 | – |
| Localization | |||
| sugar transporter ERD6-like 4 | 39.5 | 259.6 | 6.6 |
| Nucleic acid metabolic process | |||
| Superkiller viralicidic activity 2-like 2 | 259.5 | 868.1 | 3.3 |
| Organic substance metabolic process | |||
| plastid alpha-1,4-glucan phosphorylase | 10.8 | 48.9 | 4.5 |
| Oxidation–reduction process | |||
| premnaspirodiene oxygenase-like | 0 | 1048.3 | – |
| Protein metabolic process | |||
| deSI-like protein sdu1 | 40.9 | 185.7 | 4.5 |
| Lysine-specific demethylase 8 | 20.9 | 131.2 | 6.3 |
| RING-H2 finger protein | 0 | 3545.2 | – |
| Subtilisin-chymotrypsin inhibitor-2A | 262.7 | 913.9 | 3.5 |
| Response to stimulus | |||
| disease resistance protein RGA2-like | 282.2 | 872.3 | 3.1 |
| Signal transduction | |||
| Serine/threonine-protein kinase CTR1 | 91.6 | 352.7 | 3.9 |
Among DEGs, genes homologous to circadian clock and Ca2+ signal transduction related genes were abundant. In ‘Norin61’, genes homologous to CCA1 and LHY showed the highest expression on DAP20. Their expressions were lower following seed development (Table 5). RSD32 showed lower expressions of CCA1 and LHY than that of ‘Norin61’ on DAP20 or on DAP30. However, genes homologous to TOC1 expressed higher in RSD32 than in ‘Norin61’ at all developmental stages. An homologous gene to PHYTOCLOCK1 showed higher expression in RSD32 on DAP20 and DAP30, similarly to TOC1, but the difference was less apparent on DAP40. Regarding other circadian clock related genes, genes homologous to LUX-B and CONSTANS-LIKE showed higher expressions in RSD32. In addition, genes homologous to LNK1, FAR1, and RVE6-LIKE showed lower expression in RSD32. Consequently, RSD32 mutation was inferred to affect the expressions of circadian clock related genes in different manners. DEGs related to circadian clock regulation were divided into two groups based on mutant effects on expression for inhibition or enhancement.
| Putative function | ‘Norin61’ | RSD32 | RSD32 Effect | ||||
|---|---|---|---|---|---|---|---|
| DAP20 | DAP30 | DAP40 | DAP20 | DAP30 | DAP40 | ||
| Circadian rhythm | |||||||
| CCA1 | 1407.7 | 747.8 | 435.2 | 313.7 | 138.7 | 686.5 | – |
| LHY | 139.5 | 117.4 | 39.6 | 27.2 | 18.2 | 43.9 | – |
| TOC1/PPR1 | 251.2 | 496.3 | 532.0 | 594.9 | 714.8 | 694.3 | + |
| PHYTOCLOCK 1 | 3.0 | 161.1 | 557.0 | 70.6 | 536.4 | 448.6 | + |
| LUX-B | 0.0 | 68.5 | 134.9 | 30.9 | 246.9 | 107.0 | + |
| LNK1 | 902.4 | 899.7 | 414.4 | 244.4 | 432.2 | 480.1 | – |
| FAR1-related sequence 5-like | 1.4 | 617.8 | 164.2 | 0.0 | 176.2 | 86.6 | – |
| RVE6-like | 241.9 | 110.0 | 9.2 | 48.5 | 38.4 | 27.3 | – |
| CONSTANS-like | 55.1 | 172.8 | 282.5 | 162.8 | 458.4 | 317.4 | + |
| Ca signaling | |||||||
| calcium-dependent protein kinase | 1010.3 | 184.9 | 93.9 | 184.9 | 148.4 | 122.7 | – |
| calcium-binding protein | 93.6 | 6.1 | 0.0 | 0.0 | 20.2 | 4.7 | – |
| calmodulin-related protein | 581.5 | 93.8 | 2.4 | 133.8 | 77.7 | 7.1 | – |
| calmodulin-binding protein 60 D-like | 213.9 | 46.8 | 5.9 | 46.0 | 62.2 | 8.1 | – |
| CBL-interacting protein kinase 31 | 358.5 | 69.4 | 7.6 | 103.7 | 41.4 | 5.9 | – |
| calmodulin-binding receptor-like cytoplasmic kinase 3 | 114.2 | 35.4 | 11.9 | 33.2 | 27.0 | 11.0 | – |
| EF-hand Ca2+-binding protein CCD1 | 334.6 | 47.9 | 2.3 | 81.5 | 71.8 | 2.3 | – |
Effects of RSD32 on gene expression represent + (positive) and – (negative).
In ‘Norin61’, genes homologous to calcium signaling pathway related genes, CALMODULIN-BINDING RECEPTOR LIKE CYTOPLASMIC KINASE 3, CALCIUM-BINDING PROTEIN, CALMODULIN-RELATED PROTEIN, CBL-INTERACTING PROTEIN KINASE 31, CALMODULIN-BINDING PROTEIN 60D-LIKE, and CALCIUM-DEPENDENT PROTEIN KINASE were found to be expressed specifically on DAP20. Their expressions were found to be diminished on DAP30 and DAP40 (Table 4). These genes showed specific expressions at the middle developmental stage. Expressions of genes homologous to calcium signaling pathway related genes were found to be markedly inhibited on DAP20 in RSD32. All Ca signaling pathway related genes were similarly down-regulated.
‘Norin61’ is a pre-harvest sprouting-tolerant cultivar with strong seed dormancy. Although seed dormancy was maintained until DAP50, dormancy release was found on DAP60 and in later developmental stages. By contrast, RSD32 showed reduced seed dormancy on DAP40. Dormancy was found to be completely broken on DAP50. Degrees of seed dormancy in ‘Norin61’ and RSD32 differed at the late developmental stages (DAP40 and DAP50). Both lines showed low germination ability in whole seeds. No difference was observed at middle developmental stages (DAP20 and DAP30). Because half seeds, which have been released from dormancy, show poor germination, germination ability is not fully developed at this stage. Transcriptome analysis of gene expression in embryos of ‘Norin61’ and RSD32 at different developmental stages revealed conspicuously different gene expression in these lines at middle developmental stages, but not at late developmental stages, which attests to their different degrees of seed dormancy. These results suggest that RSD32 expresses at the middle developmental stage or at an even earlier stage before seed dormancy development. Shallower seed dormancy in RSD32 is associated with genes expressed at the middle developmental stage.
Genes homologous to circadian clock regulation related genes are differentially expressed in embryos of ‘Norin61’ and RSD32 at the middle developmental stages. For the component of central oscillator, genes homologous to CCA1 and LHY were down-regulated in RSD32. However, TOC1 and PHYTOCLOCK1 were up-regulated in RSD32. In Arabidopsis, CCA1 and LHY are the morning-expressed type; TOC1 and PHYTOCLOCK1 are the evening-expressed type (Seung et al. 2012). Apparently, RSD32 affects the expression of circadian clock regulation related genes depending on the circadian clock regulation function. Moreover, genes homologous to LNK1, FAR1-RELATED SEQUENCE5-LIKE, RVE6-LIKE, and CONSTANS-LIKE, which interact with clock components, showed modified expression in RSD32. For Arabidopsis, Penfield and Hall (2009) reported that circadian clock related genes are involved in dormancy release and that they affect the response to ABA and gibberellic acid (GA). Footitt et al. (2017) reported that the balance between the evening and morning phases of the clock contributes to the interpretation of temperature signals, thereby determining cycles of dormancy induction and relief in Arabidopsis. Aberrant functioning of central oscillation affects ABA biosynthesis, signal transduction, and several abiotic stress tolerances (Adams et al. 2018, Fornara et al. 2015, Kim et al. 2013b, Kolmos et al. 2014, Lee et al. 2016, Miyazaki et al. 2015, Nakamichi et al. 2012, Sanchez-Villarreal et al. 2013, Seung et al. 2012). The circadian clock might regulate several stress responses through ABA biosynthesis and the signal transduction pathway. Although the relation between circadian clock regulation and seed dormancy remains unknown in wheat, the reduction of seed dormancy in RSD32 might result from aberrant ABA signaling derived from irrelevant regulation of the circadian clock.
Genes homologous to calcium signaling pathway related genes were down-regulated in RSD32 embryos. In actuality, Ca2+ signal transduction is involved in several stress responses. In this study, expressions of Ca2+ signaling genes were found to be lower in embryos of RSD32. Irregular Ca2+ signaling induces inappropriate responses to environmental stresses. However, RSD32 showed no growth defect. RSD32 mutation might limit the effects of irregular Ca2+ signaling in the physiological traits of seed in a seed-specific manner.
This study identified Ca2+ sensor proteins, calmodulin-related protein, CDPK and CIPK as down-regulated genes in RSD32. In Arabidopsis, Ca2+ influx, and the expressions of CALMODULIN-LIKE PROTEIN 39 (CML39), CALCIUM DEPENDENT PROTEIN KINASE (CDPK, CPK) and CBL-INTERACTING PROTEIN KINASE (CIPK) affect ABA signaling (Edel and Kudla 2016, Kong et al. 2015, Midhat et al. 2018, Sanyal et al. 2017, Zhao et al. 2011, Zhou et al. 2015). In monocot species, CIPK and CPK also affect sensitivity to ABA (Chen et al. 2017, Jiang et al. 2013, Wang et al. 2018). Furthermore, Somyong et al. (2011, 2014) reported that region-located wheat pre-harvest sprouting regulating QTL, QPhs.cnl-2B.1, involved several genes associated with Ca2+ signaling pathway, such as CDPKs and CALMODULIN/Ca2+-DEPENDENT PROTEIN KINASE. This study found genes homologous to calcium signaling pathway related genes to be temporarily expressed in wheat embryos on DAP20: the middle developmental stage. Temporal induction of these genes was lost in RSD32. Seed dormancy induction might be disturbed by attenuated Ca signaling. Martí Ruiz et al. (2018) reported that CALMODULIN-LIKE 24 (CML24) regulates the expression of TOC1 through Ca2+-dependent pathway in Arabidopsis. Calcium signaling might affect the expression of circadian clock related genes in wheat. Relations among seed dormancy, ABA signal transduction, circadian clock regulation and Ca2+ signaling remain unknown, but they should be investigated further, especially for wheat. Few reports have described studies of the functions of circadian clock and Ca2+ signaling on the regulation of wheat seed dormancy. Results of the present study show that RSD32 is a useful tool for investigating the complex network of these regulatory pathways in wheat. Genes showing diverse functions other than circadian clock and Ca2+ signal transduction, such as the gene expression, biological and metabolic processes, are also involved in DEGs. Their effects on seed dormancy are unknown. Relations among these DEGs, circadian clock, Ca2+ signals and seed dormancy should be investigated.
Wheat embryos observed at DAP30 appear to be fully differentiated. Furthermore, the fresh and dry weights of endosperm reach their respective maximum values by DAP30 (Noda et al. 1994). Seed development is completed at the middle developmental stage (DAP15–DAP30). Moisture contents of seeds remain high at this stage. During the late developmental stage (DAP30–DAP50), moisture contents of seeds decrease; seeds enter the dormant state. Noda et al. (1994) reported that reserve accumulation in endosperm and the dry weight increase of embryos between DAP10 and DAP30 as a physiologically distinctive phase. Physiological states differ between seeds in their middle and late developmental stages. Although some DEGs are found to be down-regulated in RSD32 at DAP20 and DAP30, most DEGs identified on DAP40 are specifically expressed. No overlap was observed with other developmental stages. These results also indicate that seeds on DAP20 and DAP30 have similar physiological conditions, but they differ from those found on DAP40. Most studies investigating the regulation of seed dormancy have specifically examined the regulatory pathways of the late developmental stage. In monocot species, MFT, MAP KINASE KINASE and AlaAT have been identified as QTLs for regulating seed dormancy (Nakamura et al. 2011, Sato et al. 2016, Torada et al. 2016). These genes are associated with maintenance and release of seed dormancy. They express at the late developmental stage. Because RSD32 expresses at the middle developmental stage, RSD32 might be an important gene for regulating seed dormancy, acting more upstream in the regulation pathway. In Arabidopsis, DOG1 and seed maturation regulators function to regulate seed dormancy and express at early to middle developmental stages (Bentsink et al. 2006, Giraudat et al. 1992, Kagaya et al. 2005, Kroj et al. 2003, Lotan et al. 1998, Luerßen et al. 1998, Stone et al. 2001, To et al. 2006). Wheat genes homologous to DOG1 and seed maturation regulators are also expressed at early to middle developmental stages (Rikiishi and Maekawa 2014). Rikiishi et al. (2010) reported decreased expression of TaDOG1 in the embryos of RSD32. These results suggest that RSD32 acts upstream on TaDOG1 function. Regulation factors expressed at the middle developmental stage might be associated with seed dormancy initiation. Although many studies have examined the development and maintenance of dormancy, the mechanisms regulating initiation and induction of dormancy remain unknown. Early events of seed dormancy regulation in wheat are elucidated by the identification of RSD32 function. Furthermore, understanding the relations among regulation systems expressed at different developmental stages is necessary to elucidate the overall network regulating seed dormancy.
Conceived and designed the experiments: KR, MS, MM. Performed the experiments: KR. Analyzed the data: KR, MS, MM. Contributed reagents/materials/analysis tools: KR, MS, MM. Wrote the paper: KR. All authors read the manuscript and agreed to its submission.
This research was supported by the Ohara Foundation for Agricultural Science and by the Elizabeth Arnold Fuji Foundation.